

Background: While Electronic Medical Records (EMR) constitute a rich resource for research into various diseases, their unstructured format often poses practical challenges. For instance, retrieval of the records belonging to all patients with a particular outcome is often accomplished with naïve methods such as exact word matching. A more advanced alternative is to employ methods of Machine Learning (ML) for text classification. Rather than requiring a set of rules, an ML-model extracts these rules by itself given sufficient example records with known annotations.
Objectives: To build a reliable classifier with machine learning techniques that can identify Rheumatoid Arthritis (RA) cases in provided EMR entries.
Methods: Data was acquired from the HiX-EMR database consisting of 2,771 patients that visited the rheumatology outpatient clinic of the Leiden University Medical Centre between 2007 and 2018. This database featured a total of 38,216 entries. The first visit entry (if available) was selected per patient for annotation, resulting in a total of 1,361 entries. The annotated sample was then randomly split into an equally sized training and test set. Both sets were preprocessed and then classified with the following methods: Exact word-matching, Naive Bayes (NB), Decision Tree, Gradient Boosting (GB), Neural Networks and Support Vector Machines (SVM), see
Finally, the performance of the models was evaluated with a receiver operating characteristic (ROC) curve analysis via the pROC R-package [2]. The Delong test was used to assess the 95% confidence intervals (CI) and to determine the difference in performance between the word-matching method and the ML-models.
Results: The exact word-matching approach resulted in an area under the curve (AUC) of 0.76 (CI: 0.7265-0.7783), see figure. Likewise, the ML-models resulted in relatively high AUC-scores (CI) as well: NB =0.83 (0.80-0.86), SVM=0.91 (0.89-0.93), Neural Networks=0.92 (0.90-0.94) and the GB-method with a 0.94 (0.92-0.96). The Decision Tree showed the worst performance with an AUC-ROC of only 0.51 (0.49-0.56). In comparison to the exact word-matching ROC-curve, all the ML-models showed a significant difference: Decision Tree (p<2.2e-16), NB (p= 0.004), Neural Networks (p<2.2e-16), GB (p<2.2e-16) and the SVM (p=4.0e-16).
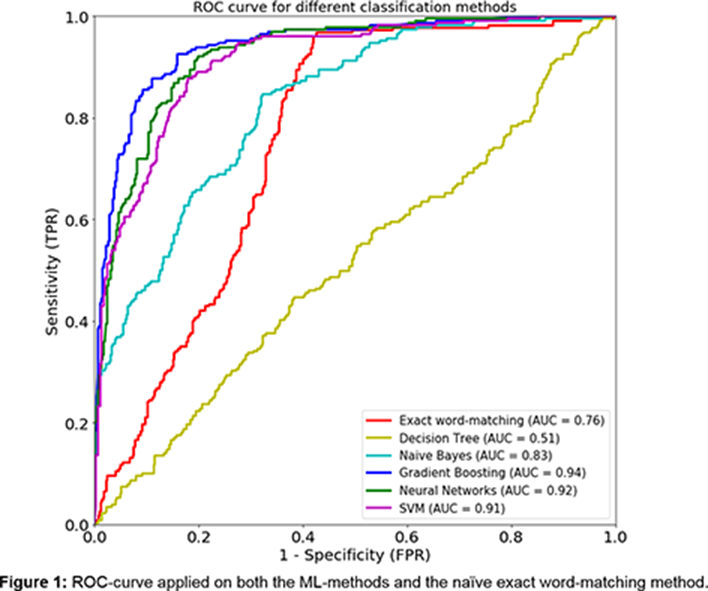
Conclusion: The Gradient Boosting, Neural Networks, SVM and Naïve Bayes models all showcased a significantly better performance than a naïve exact word matching, which establishes these ML-methods as an efficient approach for data extraction from EMR.
REFERENCES:
[1] Pedregosa, F. et al. JMLR (2011) 12: 2825-2830
[2] Robin, X. et al. BMC Bioinformatics. (2011) 12: 77
Disclosure of Interests: Tjardo Maarseveen: None declared, Thomas Huizinga Consultant for: Merck, UCB, Bristol Myers Squibb, Biotest AG, Pfizer, GSK, Novartis, Roche, Sanofi-Aventis, Abbott, Crescendo Bioscience Inc., Nycomed, Boeringher, Takeda, Zydus, Epirus, Eli Lilly, Marcel Reinders: None declared, Erik van den Akker: None declared, Rachel Knevel: None declared
DOI: 10.1136/annrheumdis-2019-eular.2408