

Background: Up to now, three groups used single cell RNA sequencing (scRNA-seq) to analyse the synovium in arthritis using different methods and material to measure RNA expression on a single cell level: Ref. 1 used unsorted dissociated synovial cells and a droplet based method; Refs 2 and 3 performed scRNA-seq on sorted cell populations.
Objectives: The aim of this study was to perform a meta-analysis of scRNA-seq data of the synovium in arthritis: 1) to define synovial fibroblast (SF) phenotypes, 2) to confirm differences across SF clusters between rheumatoid arthritis (RA) and osteoarthritis (OA) and 3) to analyse joint specific differences between SF phenotypes.
Methods: In addition to the available count matrices [1-3], we used unsorted dissociated synovial cells from three patients with undifferentiated arthritis (UA) with a droplet-based method (10x Genomics). We followed a standard protocol [4] to integrate the datasets into a shared space, even in the presence of extensive technical and/or biological differences (“batch-corrected”). SF were selected as previously described ( PDPN+ , ISLR+ , COL1A2+ , PTPRC -) [1-3]. We used a minimum log2 FC of 0.25 for average expression of genes in a cluster relative to the average expression in all other clusters combined to define marker genes. R with Seurat, Monocle and clusterProfiler packages were used for scRNA-seq analysis, pseudotime trajectory analysis and pathway enrichement analysis, respectively. Quantitative PCR (qPCR) (n=6-14 per location and disease), immunohistochemistry (IHC) and Krenn synovitis score (n=5-15 per location and disease) were performed according to standard protocols.
Results: Data from 29 RA, 3 UA and 6 OA patients were analysed. From a total of 29’448 cells, we identified 14’787 (50%) with a fibroblast phenotype. Of those, we determined 5 subpopulations (
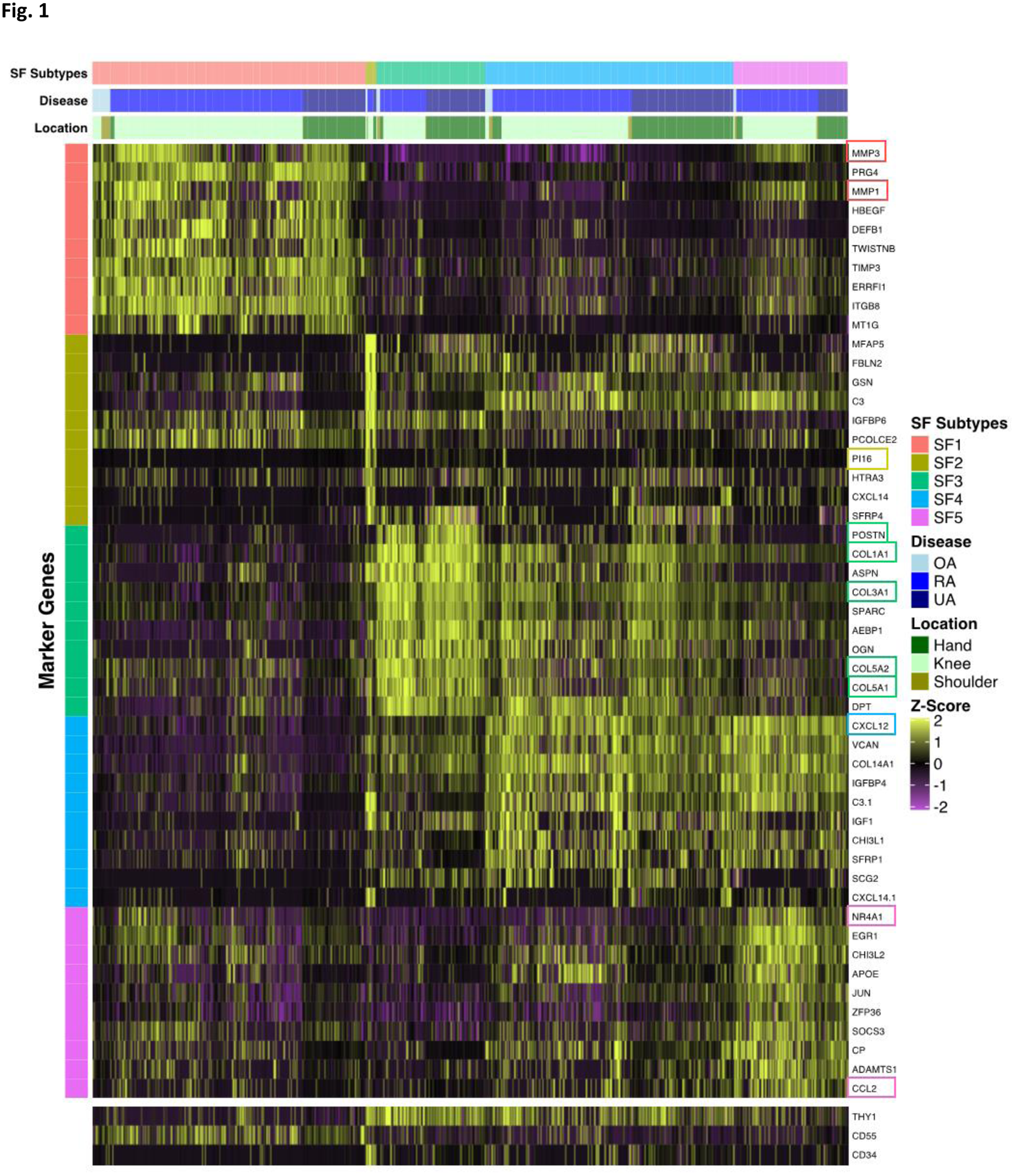
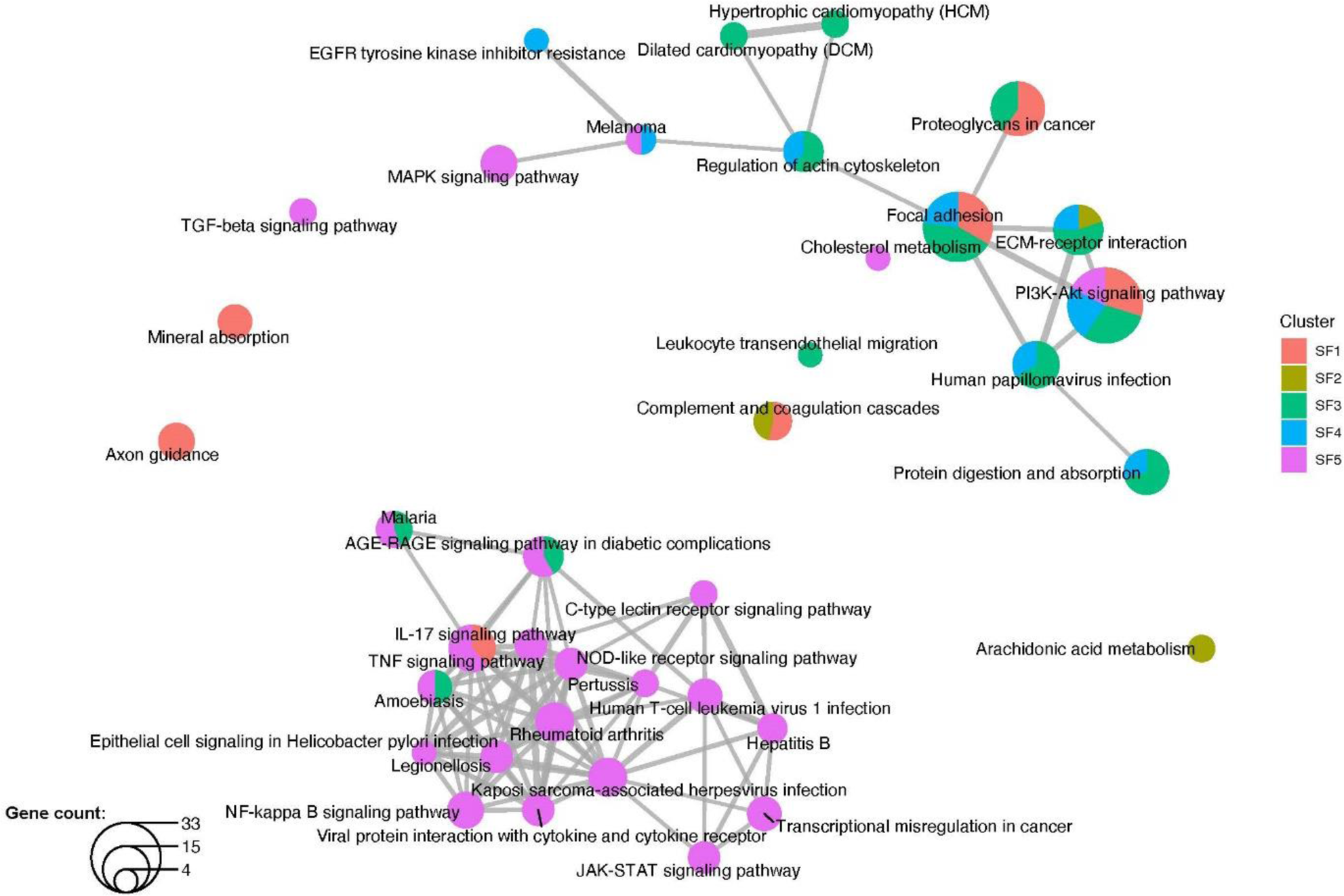
Graph 1
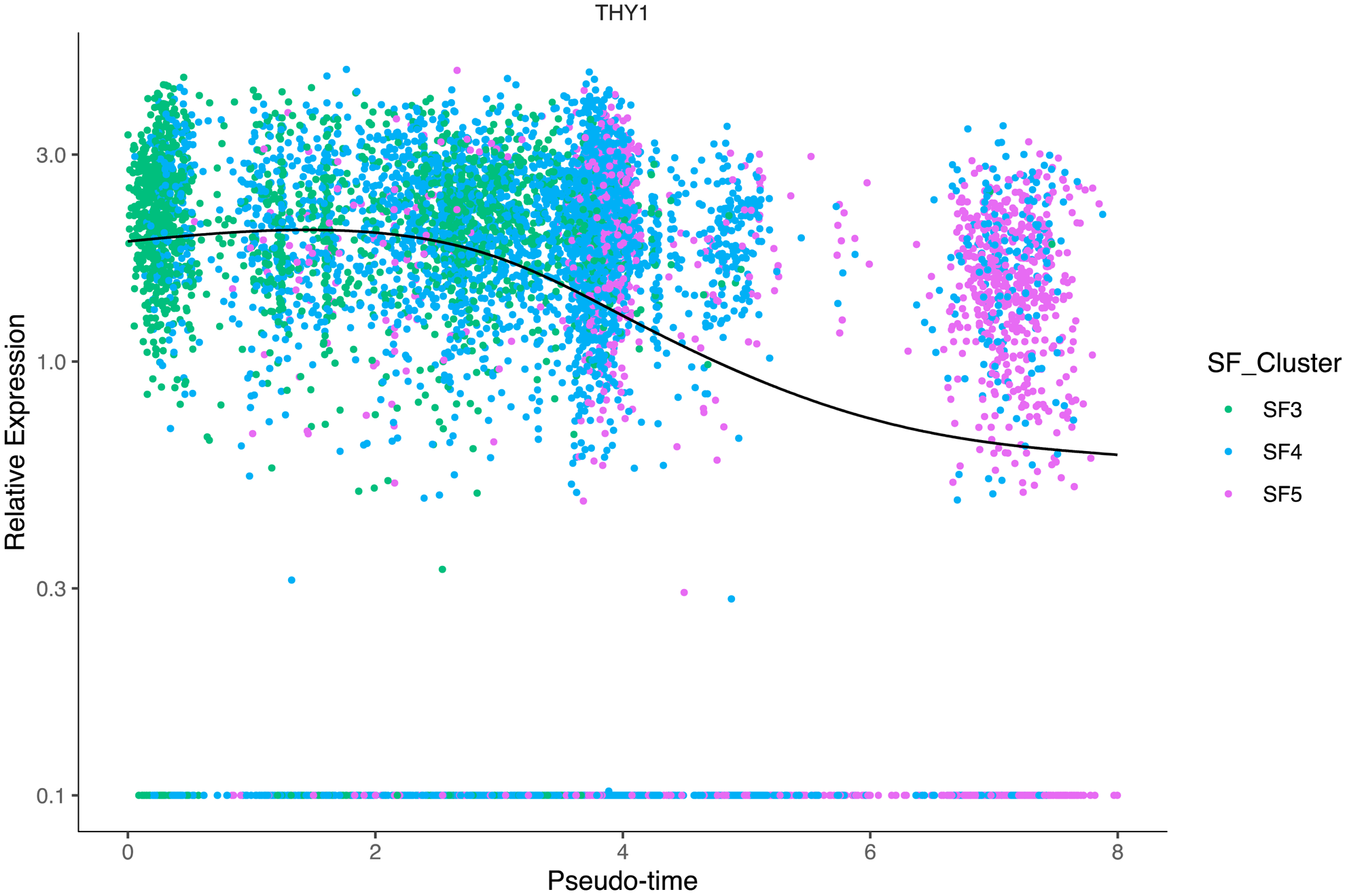
Conclusion: In our meta-analysis, we found comparable subtypes of fibroblasts as in the individual analyses [1-3], showing the robustness of cell phenotype identification using scRNA-seq. The different SF phenotypes appear to be plastic cell states rather than fixed cell subtypes, whose development is controlled by an interrelation between pathological changes in the synovium and joint location.
REFERENCES:
[1]Stephenson et al. Nat. Commun. 2018
[2]Mizoguchi et al. Nat. Commun. 2018
[3]Zhang et al. Nat Immunol. 2019
[4]Stuart, Butler, et al. Cell 2019
Disclosure of Interests: Raphael Micheroli: None declared, Mojca Frank-Bertoncelj: None declared, Sam G. Edalat: None declared, Kerstin Klein: None declared, Tadeja Kuret: None declared, Kristina Buerki: None declared, Adrian Ciurea Consultant of: Consulting and/or speaking fees from AbbVie, Bristol-Myers Squibb, Celgene, Eli Lilly, Merck Sharp & Dohme, Novartis and Pfizer., Oliver Distler Grant/research support from: Grants/Research support from Actelion, Bayer, Boehringer Ingelheim, Competitive Drug Development International Ltd. and Mitsubishi Tanabe; he also holds the issued Patent on mir-29 for the treatment of systemic sclerosis (US8247389, EP2331143)., Consultant of: Consultancy fees from Actelion, Acceleron Pharma, AnaMar, Bayer, Baecon Discovery, Blade Therapeutics, Boehringer, CSL Behring, Catenion, ChemomAb, Curzion Pharmaceuticals, Ergonex, Galapagos NV, GSK, Glenmark Pharmaceuticals, Inventiva, Italfarmaco, iQvia, medac, Medscape, Mitsubishi Tanabe Pharma, MSD, Roche, Sanofi and UCB, Speakers bureau: Speaker fees from Actelion, Bayer, Boehringer Ingelheim, Medscape, Pfizer and Roche, Caroline Ospelt Consultant of: Consultancy fees from Gilead Sciences.