

Background: Rheumatoid arthritis (RA) is a chronic, inflammatory synovitis based systemic disease of unknown etiology 1 . The genes and pathways in the inflamed synovium of RA patients are poorly understood.
Objectives: This study aims to identify differentially expressed genes (DEGs) associated with the progression of synovitis in RA using bioinformatics analysis and explore its pathogenesis 2 .
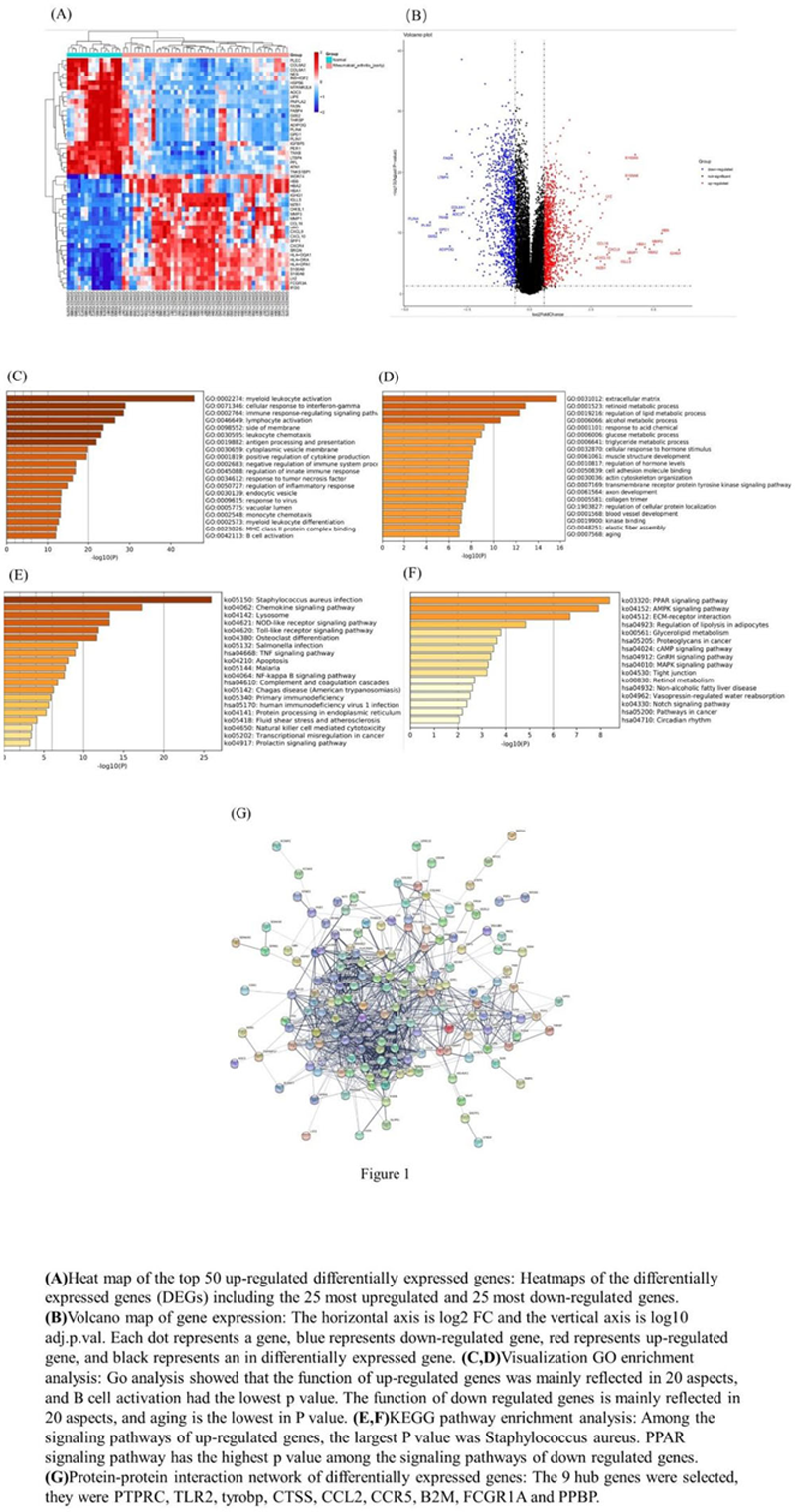
Methods: RA expression profile microarray data GSE89408 were acquired from the public gene chip database (GEO), including 152 synovial tissue samples from RA and 28 healthy synovial tissue samples. The DEGs of RA synovial tissues were screened by adopting the R software. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed. Protein-protein interaction (PPI) networks were assembled with Cytoscape software.
Results: A total of 654 DEGs (268 up-regulated genes and 386 down-regulated genes) were obtained by the differential analysis. The GO enrichment results showed that the up-regulated genes were significantly enriched in the biological processes of myeloid leukocyte activation, cellular response to interferon-gamma and immune response-regulating signaling pathway, and the down-regulated genes were significantly enriched in the biological processes of extracellular matrix, retinoid metabolic process and regulation of lipid metabolic process. The KEGG annotation showed the up-regulated genes mainly participated in the staphylococcus aureus infection, chemokine signaling pathway, lysosome signaling pathway and the down-regulated genes mainly participated in the PPAR signaling pathway, AMPK signaling pathway, ECM-receptor interaction and so on. The 9 hub genes (PTPRC, TLR2, tyrobp, CTSS, CCL2, CCR5, B2M, fcgr1a and PPBP) were obtained based on the String database model by using the Cytoscape software and cytoHubba plugin 3 .
Conclusion: The findings identified the molecular mechanisms and the key hub genes of pathogenesis and progression of RA.
REFERENCES:
[1]Xiong Y, Mi BB, Liu MF, et al. Bioinformatics Analysis and Identification of Genes and Molecular Pathways Involved in Synovial Inflammation in Rheumatoid Arthritis. Med Sci Monit 2019;25:2246-56. doi: 10.12659/MSM.915451 [published Online First: 2019/03/28]
[2]Mun S, Lee J, Park A, et al. Proteomics Approach for the Discovery of Rheumatoid Arthritis Biomarkers Using Mass Spectrometry. Int J Mol Sci 2019;20(18) doi: 10.3390/ijms20184368 [published Online First: 2019/09/08]
[3]Zhu N, Hou J, Wu Y, et al. Identification of key genes in rheumatoid arthritis and osteoarthritis based on bioinformatics analysis. Medicine (Baltimore) 2018;97(22):e10997. doi: 10.1097/MD.0000000000010997 [published Online First: 2018/06/01]
Acknowledgements: This project was supported by National Science Foundation of China (82001740), Open Fund from the Key Laboratory of Cellular Physiology (Shanxi Medical University) (KLCP2019) and Innovation Plan for Postgraduate Education in Shanxi Province (2020BY078).
Disclosure of Interests: None declared