

Background: Systemic mastocytosis (SM) is a rare monoclonal mast cell disease, associated with vertebral osteoporosis (OP) in ~30% of the patients. This OP frequently leads to multiple vertebral fractures, in particular in young or premenopausal patients. Soluble mediators secreted by the pathogenic mast cells and subsequent activation of osteoclasts are thought to explain the development of such OP. However, data about the nature and the regulation of the pro-osteoporotic factors is limited. We hypothesize that a transcriptomic approach, assessing bone marrow cells involved in this pathological condition, can identify the specific determinants of fragility fractures in SM.
Objectives: To identify a bone marrow transcriptomic profile associated with osteoporosis in SM.
Methods: We analysed clinical data and bone marrow samples collected at diagnosis in SM patients from our Reference Centre for Mastocytosis (CEREMAST) and fulfilling the SM WHO criteria. All patients signed an informed consent. Twelve SM patients with OP and fragility fracture (OP group) were age- and gender-matched with 12 SM patients without OP or fragility fracture (non-OP group). We used a Nanostring nCounter approach to compare the bone marrow mRNA profile. We used a predefined panel of 800 transcripts relevant for an osteo-immunological analysis. Genes expressions were normalized and compared by a Wilcoxon test with R software. We performed a pathway analysis with C5, C7, KEGG and Reactome databases.
Results: Both OP and non-OP groups included 8 women and 4 men, with similar average age (OP 56.1 +/- SD 8.7 vs non-OP 57.1 +/- 9.3). The 12 patients from the OP group had at least 1 vertebral fracture (VF) with an average number of VF of 4.9 +/- 2.6. Lumbar and hip bone mineral density was significantly lower in the OP group. The transcriptomic analysis revealed a specific profile associated with osteoporotic vertebral fractures in SM, with 26 differentially expressed genes (see
Heatmap of the differentially expressed genes in bone marrow from SM patients with osteoporosis and fragility fracture versus without osteoporosis
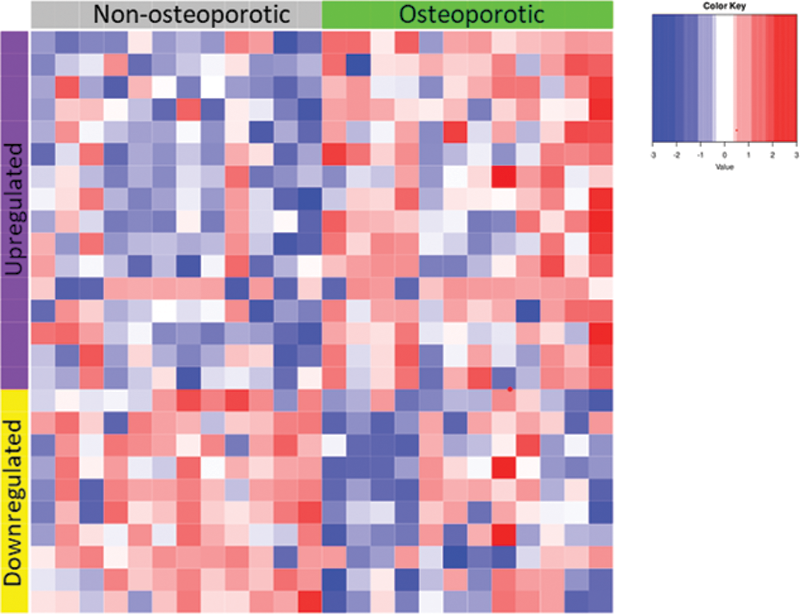
Conclusion: Our results highlight that a transcriptomic analysis of bone marrow is relevant to identify biomarkers associated with OP in SM. In addition to the pathogenic role of clonal mast cells and of monocytic lineage, this condition may involve non-clonal immune cells such as T cells.
Acknowledgements: This work has been supported by the GRIO (Groupe Français de Recherche et d’Infrmation sur les Ostéoporoses)
Disclosure of Interests: Yannick Degboe Grant/research support from: This work has been supported by a Novartis grant, Arnaud Constantin: None declared, Adeline Ruyssen-Witrand: None declared, Carle Paul: None declared, Patrice Dubreuil: None declared, Cristina Bulai Livideanu: None declared