

Background Despite advances in the understanding of juvenile idiopathic arthritis (JIA) pathophysiology, personalized treatments informed by gene transcriptomic profiles remain elusive.
Objectives To examine the relationship between changes in gene expression and treatment response in patients with JIA treated with tofacitinib.
Methods Whole blood samples from patients with JIA were collected using Paxgene tubes at baseline (BL) and again at week (wk) 18 of treatment with tofacitinib as part of a clinical trial (NCT02592434) [1]. Patients were classified as treatment responders (TR) if they achieved at least a JIA-American College of Rheumatology score of (JIA-ACR70) or above (NBL=47; NWK18=38); and non-responders (NR) improvement was no more than a JIA-ACR30 response (NBL=20; NWK18=8). Bulk RNA was isolated, subjected to globin/rRNA depletion, and used for generation of cDNA libraries for subsequent sequencing. RNA sequencing via Illumina Nova-seq generated 50 million reads per sample. Gene expression was compared in TR and NR (BL, wk18; change BL to wk18). Kallisto 0.48.0 was used for pseudo-quantification (GRCh38 release 94 index). Differential expression and Gene Ontology (GO) over-representation analyses were performed with dseqr 0.34.0.
Results Significant differential expression (FDR<0.05) was observed in 10,138 genes between BL and wk18. GO over-representation analysis was performed separately for up- and down-regulated genes with FDR<0.05 and absolute logFC>0.8 (n=231). For genes down-regulated with treatment (BL-wk18), ontologies related to negative regulation of gene expression, type I and type II interferon pathways were significantly over-represented (FDR<0.05), while ontologies for synapse organization and activity were over-represented among up-regulated genes (Figure 1). The latter might reflect the normalization of the immunological synapse leading to improved cytotoxic NK and T cell functions, the impairment of which has been proposed to escalate the production of IFN-γ and other proinflammatory cytokines [2]. There were no significantly differentially expressed genes (FDR < 0.05) between TR vs. NR at BL or wk18. Exploratory GO analysis in NR vs. TR at baseline (FDR<0.13, n=76 genes) suggests up-regulation of genes in ontologies related to the activation of MAP kinase activity.
Conclusion Tofacitinib treatment in JIA patients leads to widespread changes in whole blood transcriptional profiles. The most significant transcriptional changes observed included the down-regulation of genes associated with type I and type II interferon pathways, and the up-regulation of genes involved in synapse organization and activity. Potential association of MAP-kinase activation in NR at baseline needs to be further explored. MAP-kinase is one of the signalling pathways in inflammatory arthritis that can be targeted by small molecules other than JAK inhibitors. Thus, patients with a predominantly active MAP-kinase pathway could be less likely to respond to tofacitinib. If confirmed, these findings might be useful to personalize JIA treatment in the future.
References
Image/graph:
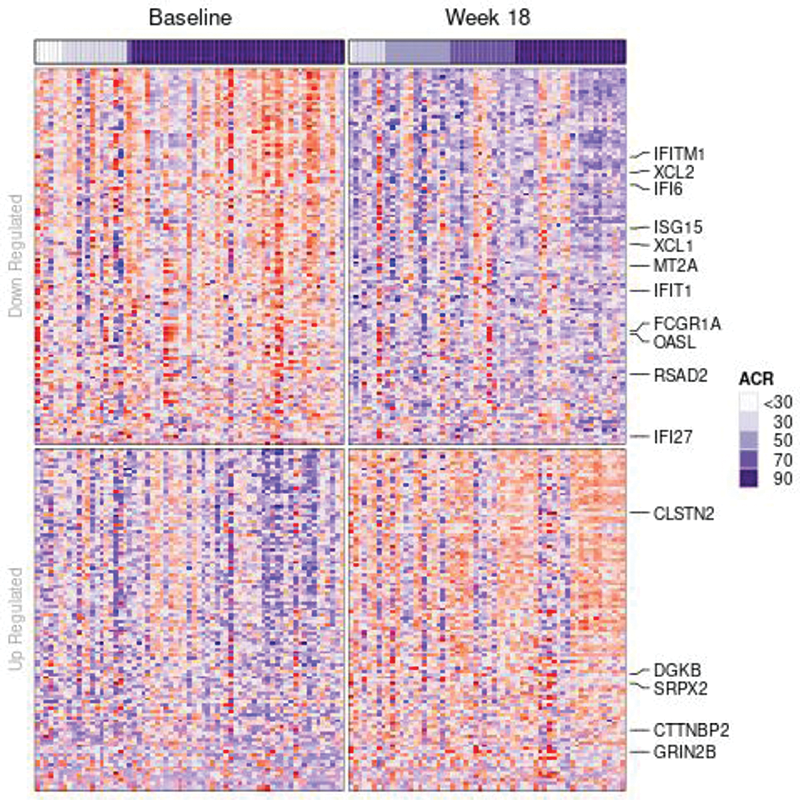
Figure 1. Heatmap showing differentially expressed genes (FDR < 0.05 and |logFC| > 1) at week 18 vs baseline in JIA patients receiving tofacitinib. Annotated genes are from ontologies related to type I and type II interferon activity (down-regulated), as well as from ontologies related to synapse organization (up-regulated).
Acknowledgements Funding: Ralph & Marian Falk Medical Trust Catalyst Award; the Cincinnati Children’s Hospital Medical Center Pediatric Musculoskeletal & Rheumatology Innovation Core center; the University of Cincinnati Center for Clinical and Translational Science and Training; Serum samples were donated by Pfizer; We thank the PRCSG and PRINTO investigators for the collection of serum samples and data used. We thank the SJIA foundation as they granted access and paid for bioinformatics services.
Role of the study sponsor: Serum samples were donated by Pfizer. Pfizer was not involved in the study design, data collection, analysis, or data interpretation. Pfizer reviewed and approved the abstract for submission.
Disclosure of Interests Esraa Eloseily: None declared, Alex Pickering: None declared, Sanjeev Dhakal: None declared, Hermine Brunner Speakers bureau: Dr. Brunner serves on the speakers’ bureau and on ad-board for Pfizer., Consultant of: Dr. Brunner is a consultant for Pfizer with funds received by CCHMC, her primary employer, Alexei Grom Consultant of: Sobi and Novartis, Grant/research support from: Sobi and Novartis, Sherry Thornton: None declared.
Keywords: bDMARD, -omics, Inflammatory arthritides
DOI: 10.1136/annrheumdis-2023-eular.680