

Background: The epidemic of coronavirus disease 2019 (COVID-19) has brought a heavy burden to the world. It is worth to note that it shares many clinical symptoms with systemic lupus erythematosus (SLE), but whether there is a similar pathological process between them is not clear.
Objectives: In this study, we analyzed the potentially similar pathogenesis between SLE and COVID-19, and explored their possible pharmacotherapy options using bioinformatics and systems biology approaches.
Methods: The common differentially expressed genes (DEGs) were extracted from the COVID-19 datasets and the SLE datasets for functional enrichment, pathway analysis and candidate drug analysis.
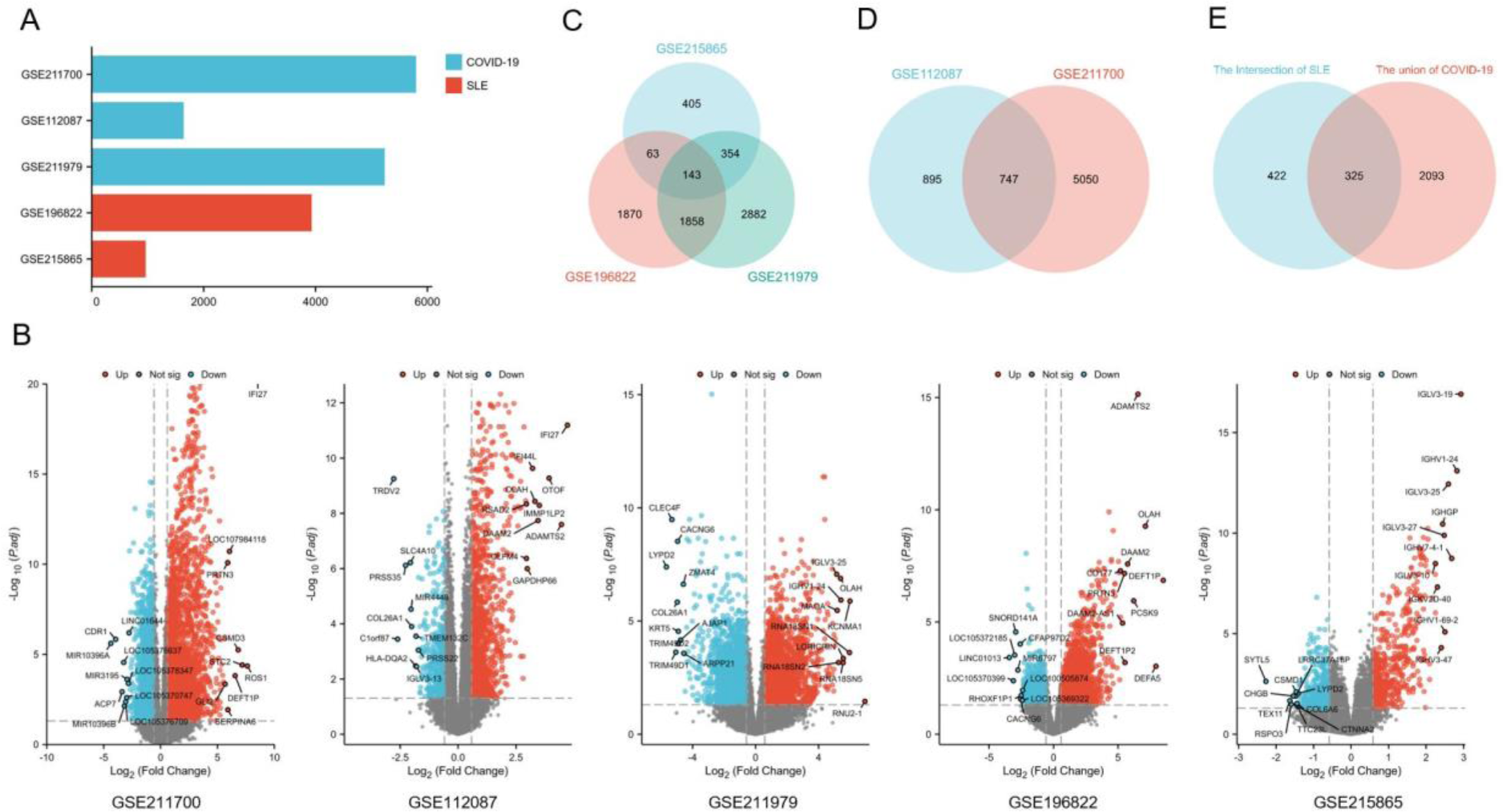
Results: Based on the two transcriptome datasets between COVID-19 and SLE, 325 common DEGs were selected. Hub genes were identified by protein-protein interaction (PPI) analysis. few found a variety of similar functional changes between COVID-19 and SLE, which may be related to the pathogenesis of COVID-19. Besides, we explored the related regulatory networks. Then, through drug target matching, we found many candidate drugs for patients with COVID-19 only or COVID-19 combined with SLE.
Conclusion: COVID-19 and SLE patients share many common hub genes, related pathways and regulatory networks. Based on these common targets, we found many potential drugs that could be used in treating patient with COVID-19 or COVID-19 combined with SLE.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.