

Background: Dermatomyositis (DM) is an inflammatory myopathy characterized by skin lesions (Gottron’s Papules) and proximal, symmetric weakness with elevation of creatin-kinase, alanine transaminase, and aspartate transaminase enzymes [1]. A higher incidence of any type of malignancy has been noted in patients with DM when compared to general population [2]. We performed an enrichment analysis on a dataset of patients with DM to investigate the pathogenesis of DM and cancer.
Objectives: This study aims to find whether genes enriched for cancer are present in muscle biopsies of patients with DM using transcriptomic data.
Methods: We downloaded the dataset GSE1551 from the GEO platform. We utilized the Gene Set Enrichment Analysis software (GSEA) to identify overrepresented genes in the set of “Pathways in cancer”. This is a curated dataset from the Kyoto Encyclopedia of Genes and Genomes (KEGG) which contains genes implicated in several pathways of different malignancies. The database for annotation, visualization and integrated discovery (DAVID) was then utilized to identify which malignancies are implicated in this cohort of patients. Protein-protein interactions were analyzed with the Molecular Complex Detection (MCODE) plugin provided by Cytoscape. Hub genes (HGs) were then obtained with the plugin Cytohubba identifying common genes in 5 networks generated by 5 different algorithms (Degree, DMNC, EcCentricity, Radiality, and Stress).
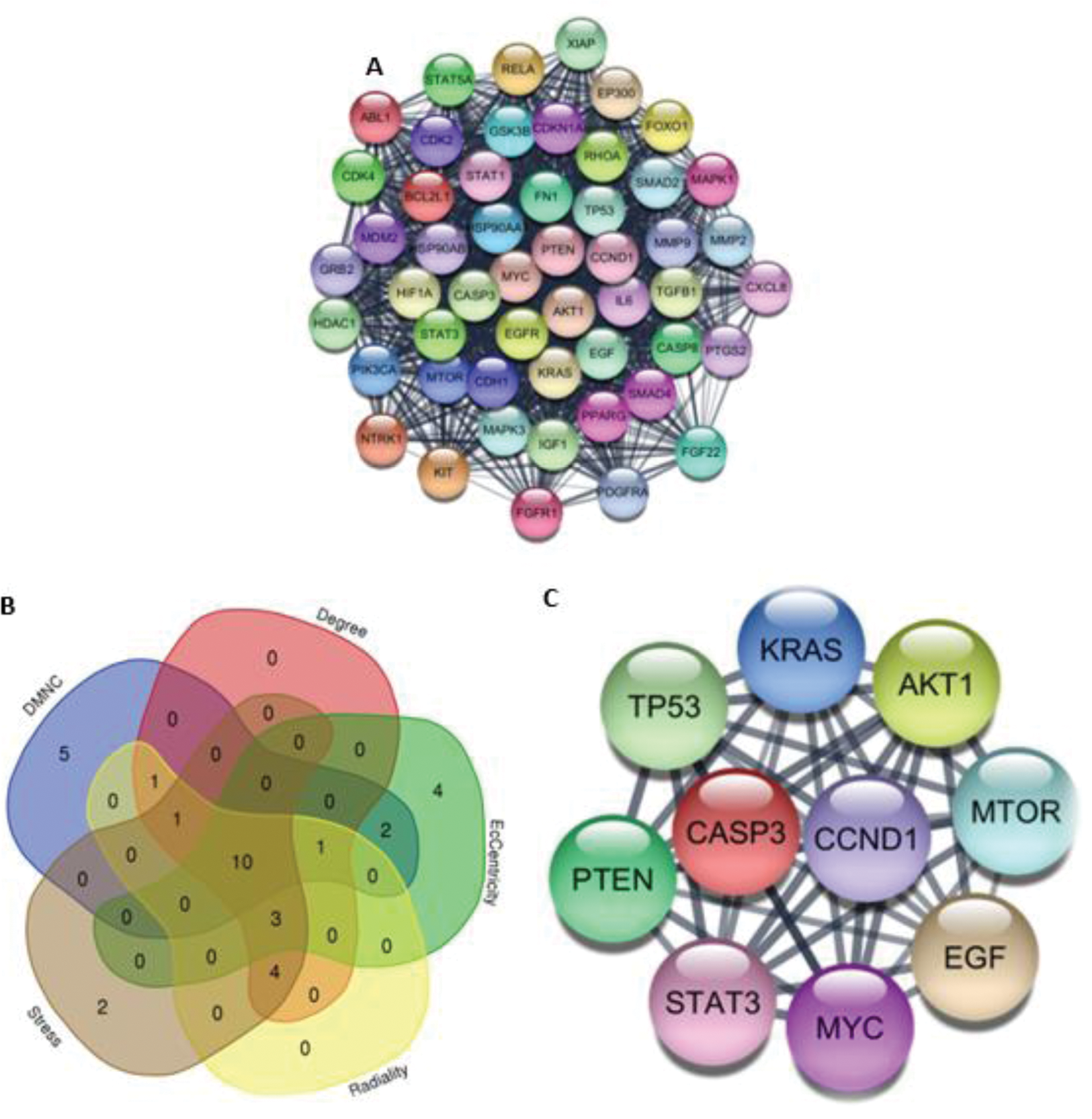
Results: 13 patients with DM were compared to 10 healthy controls. Patients of the study met the criteria for definite DM (9) and probable DM (4). 157 genes were overrepresented for pathways in cancer. These genes were found to participate in breast cancer, colorectal cancer, hepatocellular carcinoma, ovarian cancer, acute myeloid leukemia, and bladder cancer. Furthermore, intracerebral hemorrhage and Schimmelpenning Syndrome were also overrepresented in this set of genes (Table 1). 10 HGs were identified using the 5 algorithms (Figure 1).
This table shows functional annotations of the DAVID database in the category of diseases.
| Functional Annotation Analysis: Diseases | |||
|---|---|---|---|
| Category | Genes | p -Value | Benjamini |
| Breast cancer, somatic | AKT, KRAS, PIK3CA, tp53 | 2,0E-4 | 4,8E-2 |
| Colorectal cancer, somatic | AKT, EP300, NRAS, PIK3CA | 2,2E-3 | 2,6E-1 |
| Hepatocellular carcinoma, somatic | CASP8, PIK3CA, TP53 | 3,5E-3 | 2,8E-1 |
| Ovarian cancer, somatic | AKT1, CDH1, PIK3CA | 4,9E-3 | 2,9E-1 |
| Leukemia, acute myeloid, somatic | CEBPA, KIT, KRAS | 8,2E-3 | 3,9E-1 |
| Hemorrhage, intracerebral, susceptibility to | COL4A1, COL4A2 | 3,1E-2 | 1,0E0 |
| Schimmelpenning-Feuerstein-Mims syndrome, somatic mosaic | KRAS, NRAS | 4,7E-2 | 1,0E0 |
| Bladder cancer, somatic | KRAS, RB1 | 6,2E-2 | 1,0E0 |
This figure shows the PPIs of the network: A) Analysis with MCODE identified the cluster with the highest score (44.8). B) Overlapped genes using the algorithms provided by Cytohubba. C) Hub genes of malignancy within the cluster of interactions.
Conclusion: This study aimed to find if muscle biopsies of patients with DM had a transcriptional signature related to malignancy. In this analysis, we found transcriptomic evidence that a cohort of patients with DM expresses genes related to cancer in muscle biopsies. These findings are of utility to better understand the pathogenesis of malignancy and DM, as well as may allow the development of screening strategies for cancer when making a diagnosis of DM. However, further in vivo studies are warranted to corroborate these results.
REFERENCES: [1] DeWane, Madeline E et al. “Dermatomyositis: Clinical features and pathogenesis.” Journal of the American Academy of Dermatology vol. 82,2 (2020): 267-281. doi:10.1016/j.jaad.2019.06.1309.
[2] Qiang, Judy K et al. “Risk of Malignancy in Dermatomyositis and Polymyositis.” Journal of cutaneous medicine and surgery vol. 21,2 (2017): 131-136. doi:10.1177/1203475416665601.
Acknowledgements: NIL.
Disclosure of Interests: None declared.