

Background: Patients suffering from rheumatoid arthritis (RA) have been identified as vulnerable at the beginning of the COVID-19 pandemic and prioritized for vaccination programs. Patients with RA are genetically different from the general population; they have an increased carriage of shared epitope alleles, including HLA-DRB1*04:01. This allele is involved in the presentation of viral antigens (including from the spike protein of SARS-CoV-2 [1]) by antigen-presenting cell to CD4 + T cells. How genetic susceptibility to RA might influence the longevity and magnitude of the immune response to SARS-CoV-2 (natural and vaccine-induced) is unknown. One hurdle preventing studies to address this question is the lack of bioinformatics tools to track SARS-CoV-2 mutations within known HLA-DRB1*04:01-restricted immunogenic peptides. Mutations in these peptides are relevant for immune monitoring of CD4 + T cell responses against SARS-CoV-2 spike protein in patients with RA.
Objectives: Develop a bioinformatics package for mutation tracking in SARS-CoV-2 immunogenic epitopes restricted by HLA-DRB1*04:01.
Methods: Three HLA-DRB1*04:01-restricted immunogenic peptides from the Spike protein of SARS-CoV-2 were selected from the literature [1], which we refer to as S1, S2 and S3. We aligned 2,334,011 SARS-CoV-2 genomes sampled across the United Kingdom (UK) with metadata from the COVID-19 Genomics UK Consortium (COG-UK) public data resource. We developed a Python3 toolset to determine the position of each mutation and its frequency over time, including the generation of user-friendly plots to visualize mutation dynamics.
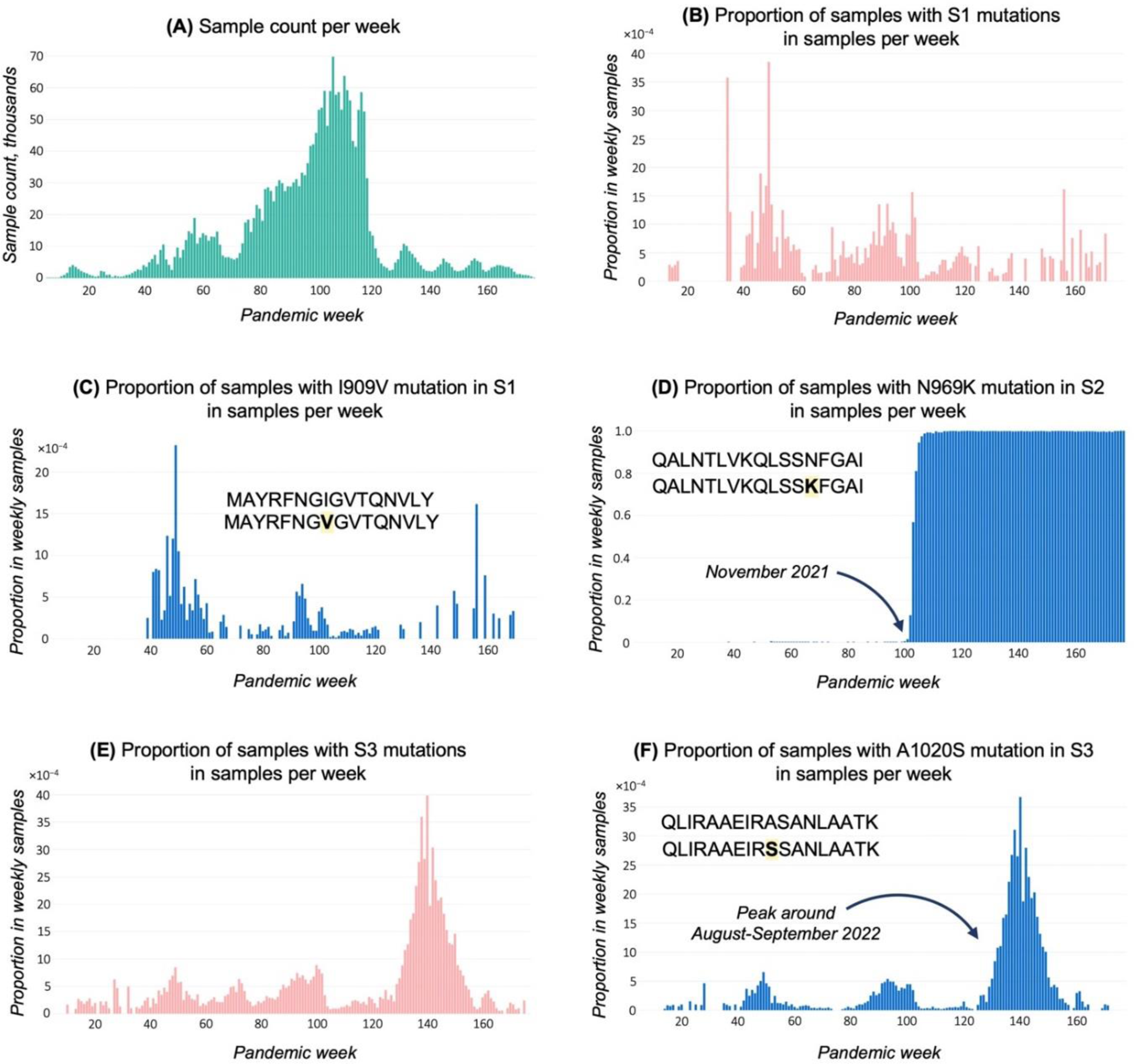
Results: We describe the development of a mutation tracker, EpitopeScan, a Python3 package with command line and graphical user interface tools facilitating the investigation of the mutation dynamics in SARS-CoV-2 epitopes via analysis of multiple sequence alignments of genomes over time. We provide an application case by examining 3 Spike protein-derived immunodominant CD4 + T cell epitopes restricted by HLA-DRB1*04:01. We show that the mutation rate in S1 and S3 is very low over the duration of the pandemic (the epitope is strongly conserved over time, Figure 1B, C, E, and F); in contrary, S2 is completely lost and replaced by a mutant with the emergence of the N969K Omicron-specific mutation in November 2021 around week 100 of the pandemic (Figure 1D). The wild-type and the mutated S2 peptides represent potential candidates to monitor variant-specific CD4 + T cell responses which were prevalent at different time points during the pandemic.
Conclusion: We have developed a SARS-CoV-2 mutation tracker which successfully manages to identify relevant mutations within CD4 + T cell epitopes restricted by the RA susceptibility allele HLA-DRB1*04:01. The peptides identified might allow to specifically track CD4 + T cell responses which emerged at different time points within the pandemic and get insights into immunological memory. Our tool will be useful to determine the longevity and magnitude of the cellular immune response against SARS-CoV-2 in RA patients.
REFERENCES: [1] Yang J, James E, Roti M, Huston L, Gebe JA, Kwok WW. Searching immunodominant epitopes prior to epidemic: HLA class II-restricted SARS-CoV spike protein epitopes in unexposed individuals. Int Immunol. 2009 Jan;21(1):63-71.
Time plots exported from EpitopeScan graphical user interface. (A) Sample count per week for analysed samples with metadata. Plots (B-F) show proportion (bound between 0 and 1) in weekly sample count for samples with: (B) S1 amino acid (AA) mutations; (C) S1 I909V mutation; (D) S2 N969K mutation; (E) S3 AA mutations; (F) S3 A1020S mutation. Proportions at plots (C, D, F) are calculated from samples with sufficient coverage and no disruptions in Spike protein open reading frame.
Acknowledgements: Disease 2019 Genomics UK Consortium (COG-UK) for providing public access to SARS-CoV-2 genome data generated with the effort of hundreds of researchers.
Disclosure of Interests: None declared.