

Background: RNA sequencing (RNA-seq) has emerged as a widely embraced technique for comprehensive gene expression profiling on a large scale. Nonetheless, there is a current absence of accessible and versatile tools that facilitate efficient exploration of RNA-seq datasets from individuals with Sjogren’s disease (SD) for researchers.
Objectives: Our goal is to create user-friendly websites based on R Shiny, designed for gene expression analyses within two distinct SD cohorts: i) an observational disease-control cohort (sicca/Sjogren’s disease), and ii) the TRACTISS randomized clinical trial, comparing Rituximab versus placebo with longitudinal data pre/post-treatment 1 .
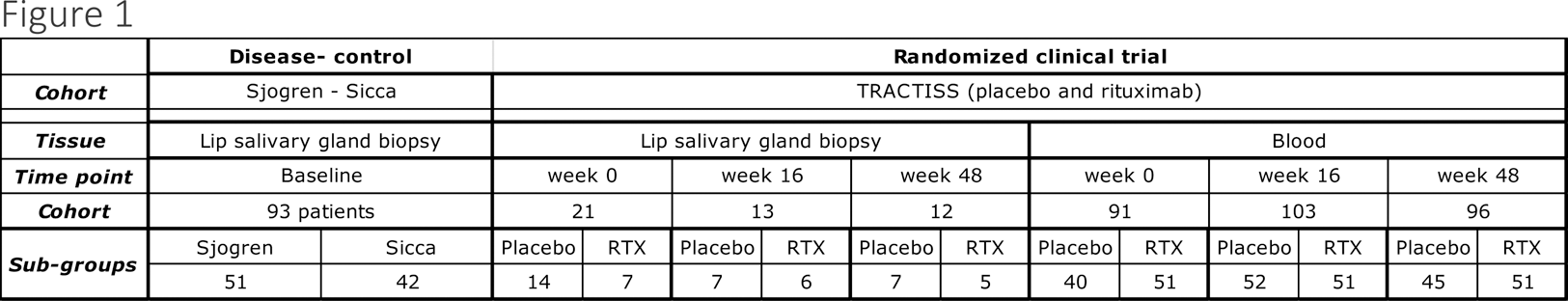
Methods: The searchable web interfaces was developed using R Shiny (v.1.7.2) to investigate the associations between individual gene transcript levels and histological as well as clinical parameters, including clinical response. These interfaces incorporate salivary gland (SG) biopsy and peripheral blood (PB) RNAseq data (Table 1) from both a disease-control (Sjogren’s/sicca) cohort and the TRACTISS randomized clinical trial, encompassing longitudinal data from Sjogren’s patients exclusively.
Results: The websites enable data exploration and visualization, facilitating the comparison of gene expression levels among defined groups (Sicca/Sjogren or Sjogren: Placebo/Rituximab) based on user-selected clinical variables. Whether the variable is continuous or categorical, users can visualize box plots or correlation plots. Additionally, correlations between individual clinical variables and all genes can be visualized, providing the list of genes significantly correlated with the variable of interest.
Differential expression analysis of RNA-seq data is available, offering interactive volcano plots that easily depict the user’s genes of interest. Users can also generate heatmaps using a custom list of genes.
For the TRACTISS randomized clinical trial, a time-series analysis of gene expression over time (with three time points for each patient) can be visualized for both SG biopsy and PB RNAseq data. Moreover, gene expression can be stratified based on response criteria, including ESSDAI improvement, CRESS, and STAR composite scores.
Conclusion: We have developed two user-friendly websites for Sjogren’s RNA-seq differential expression and time-series analysis. These intuitive platforms offer a diverse selection of interactive plots, enabling researchers to efficiently explore hypotheses, identify expression patterns, and expedite their research with prompt results. All generated results can be downloaded in a high-quality, publication-ready format.
REFERENCES: [1] Pontarini E, Sciacca E, Chowdhury F, et al. Serum and tissue biomarkers associated with CRESS and STAR response to B-cell targeted therapy in TRACTISS trial of Sjogren’s syndrome. Arthritis Rheumatol Published Online First: 10 December 2023. doi:10.1002/art.42772
Acknowledgements: NIL.
Disclosure of Interests: Elena Pontarini: None declared, Elisabetta Sciacca: None declared, Giulia Cavallaro: None declared, David Galbraith Janssen Pharmaceuticals, Alfredo Pulvirenti: None declared, Ling-Yang Hao Janssen Pharmaceuticals, Kathy Sivils Janssen Pharmaceuticals, Myles Lewis: None declared, Costantino Pitzalis: None declared, Michele Bombardieri Janssen Pharmaceuticals.