

Background: An unmet need exists to identify an ideal and reliable non-invasive biomarker that can be used to diagnose, prognosticate, and also guide the management of lupus nephritis (LN). Recent aptamer-based urine biomarker studies in LN have indicated that the urinary-activated leukocyte cell adhesion molecule (uALCAM or CD166) has one of the highest discriminatory powers to accurately predict active LN, especially in Asian and Afro-American cohorts.
Several large-scale studies in varying ethnic backgrounds are required to validate and standardize uALCAM as a biomarker of LN and to prove its reliability and reproducibility before being implemented in clinical practice. This study is our effort to assess the performance of uALCAM as a non-invasive biomarker of active lupus nephritis in a Systemic Lupus Erythematosus (SLE) cohort from Southern India.
Objectives: To estimate uALCAM in a cohort of SLE patients and assess its diagnostic performance in accurately detecting active lupus nephritis.
Methods: This is a single-centre cross-sectional study from a tertiary care centre in South India. Adults fulfilling EULAR/ACR - 2019 Criteria for the diagnosis of SLE were included in the study. The study subjects were divided into the following 3 categories: Active lupus nephritis (aLN), Active Non-Renal SLE (aNR), and Inactive Non-Renal SLE (iNR).
The disease activity of study participants was calculated according to SLEDAI 2k (Sle disease activity index- version 2k) and renal activity was calculated according to rSLEDAI (the 4 renal components of SLEDAI 2k. Sle in flare was defined as SLEDAI 2k score≥6 and SLE in remission as SLEDAI≤4. Active lupus nephritis was defined as rSLEDAI ≥ 4. uALCAM levels were measured using ELISA kits from R&D Systems Minneapolis, USA (Cat No. DY656) according to the manufacturer’s instructions. The diagnostic performance of uALCAM was assessed by plotting the receiver operating curve (ROC) analysis.
Machine Learning algorithms through Python ver. 3.8.16 and Jupyter were utilized to determine the parameters which can increase the diagnostic performance of uALCAM and identify a composite biomarker. First, the data was encoded and filtered for feature selection. Using Exploratory data analysis we plotted scatter plots and ran a subset correlation matrix to select the best parameters which increases the diagnostic accuracy of uALCAM. To determine the relationship between the target class (aLN) using the feature vectors, we initialized an XG boost classifier. Then, we trained the model with 70% input data and tested it on 30% data. The accuracy, precision, and recall achieved using a single parameter of uALCAM, and using multiple parameters were analysed to identify a composite biomarker.
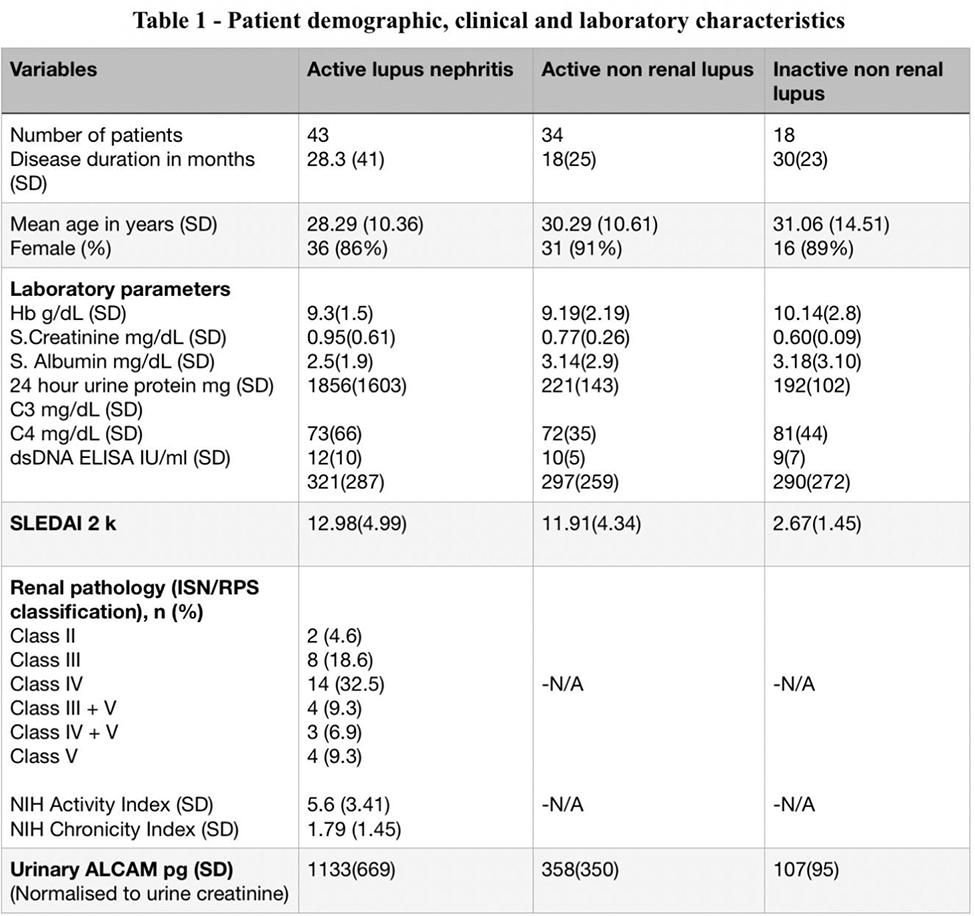
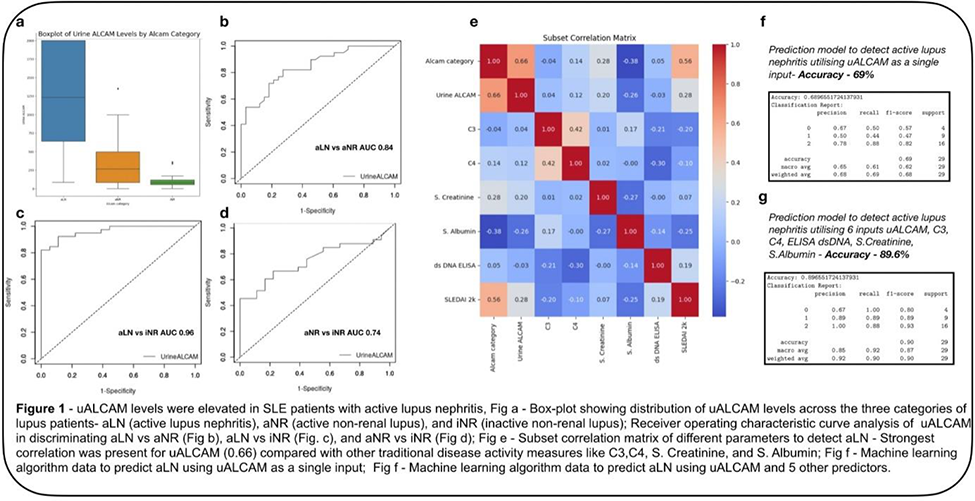
Results: We included 95 patients in the study. Table 1 provides an overview of the patient characteristics and laboratory values. The levels of uALCAM were significantly increased in aLN patients 1231 pg (IQR - 1360) when compared to aNR patients 264 pg (IQR - 414), and iNR patients 78 pg (IQR - 63), p < 0.001 (Figure 1a). ROC Analysis showed, the area under the curve (AUC) value was 0.84 (95% CI 0.75 to 0.93; p<0.001) for aLN versus aNR (Figure 1b), 0.96 (95% CI 0.92 to 1.04; p < 0.001) for aLN versus iNR (Figure 1c), and 0.74 (95% CI 0.61 to 0.87; p < 0.001) for aNR versus (Figure 1d).
The subset correlation matrix showed that uALCAM had the highest correlation with active lupus nephritis (0.66) as shown in Figure 1e. The other parameters with good correlation were SLEDAI 2k (0.56), C3 (0.42), C4 (0.42), serum creatinine (0.28), and dsDNA ELISA (0.05) and a negative correlation with S. albumin (-0.38) (Figure 1e). The accuracy of the machine learning model to predict the presence of aLN was 68% (Figure 1f) with uALCAM as a single parameter, which was boosted to 89% (Figure 1g) by including the aforementioned feature vectors.
Conclusion: Our study shows that uALCAM is an accurate biomarker to detect the presence of active lupus nephritis in a South Indian cohort of lupus patients. Through machine learning, we were able to identify other parameters like C3, C4, serum creatinine, ds DNA, and S. albumin that can be used with uALCAM as a composite biomarker to increase its accuracy.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.