

Background: The role of ROS in the development of autoimmune disease is still unclear, though some ROS mechanisms have been studied, the precise mechanisms of ROS generation in autoimmune disease, and their role in disease progression and pathogenesis is yet unknown. The role of ROS has been studied in organ-specific autoimmune diseases such as Hashimoto’s thyroiditis, inflammatory bowel disease, multiple sclerosis, and vitiligo, as well as in systemic autoimmune diseases such as rheumatoid arthritis.
Objectives: This study aims to evaluate the enrichment of molecular pathways related to human body response to chemical stress and ROS generation in primary Sjögren’s syndrome (pSS), systemic lupus erythematosus (SLE) and multiple sclerosis (MS).
Methods: A PBMC expression database for several autoimmune diseases was selected: GSE48378 for primary Sjögren’s syndrome (pSS), GSE121239 for systemic lupus erythematosus (SLE), GSE146383 for multiple sclerosis (MS) and then analyzed to explore the differentially expressed genes (DEGs) between each disease and its respective healthy control group. A total of 50 healthy control samples and 84 disease samples were obtained. GSVA was used to explore the enrichment of molecular pathways related to ROS biosynthesis and metabolism and cellular response to chemical stress. Briefly, GSVA is a method that assesses the enrichment of predefined gene sets or pathways in individual samples. The comparison of the enrichment scores calculated by GSVA for each sample between groups was done, as well as detailed gene-level analysis of altered pathways for each disease. An analytical software was used to infer relative cell populations on a given tissue from gene expression levels and correlate the cell populations of the samples with the enrichment scores for all pathways.
Results: The differences found in the enrichment scores of the studied genesets related to ROS biosynthesis, metabolism and cellular response to chemical stress were found in the datasets of pSS, SLE and MS (The statistical significance for each comparison is summarized in Figure 1). The differences in the enrichment of the studied pathways are particularly evident in pSS and SLE datasets, whereas in MS, is comparatively subtle. Several differences in cell populations were found between patient samples and healthy controls in all diseases were observed, but all the differences in the pathway enrichments could not be attributed to variations in imputed cell populations (Table 1).
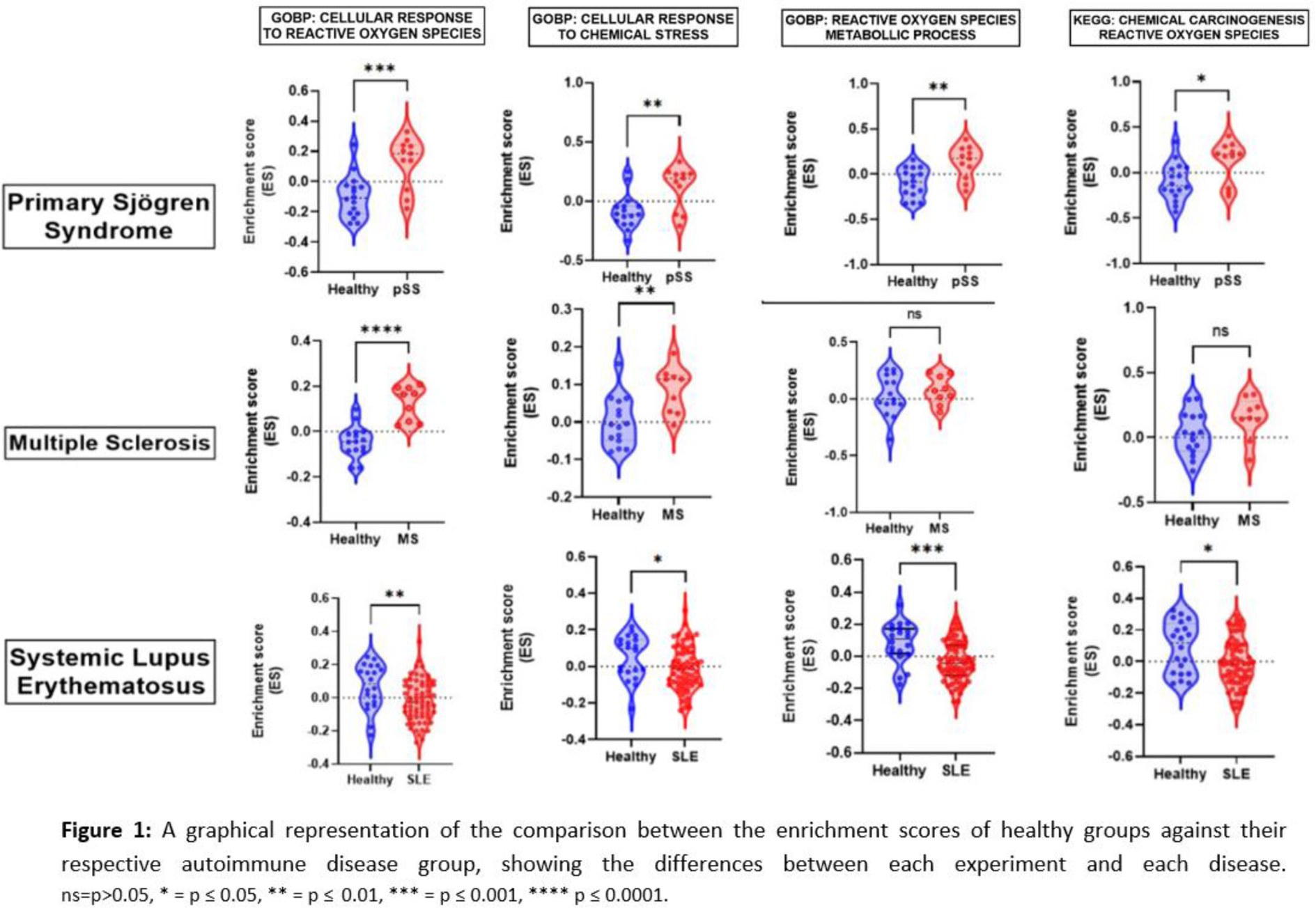
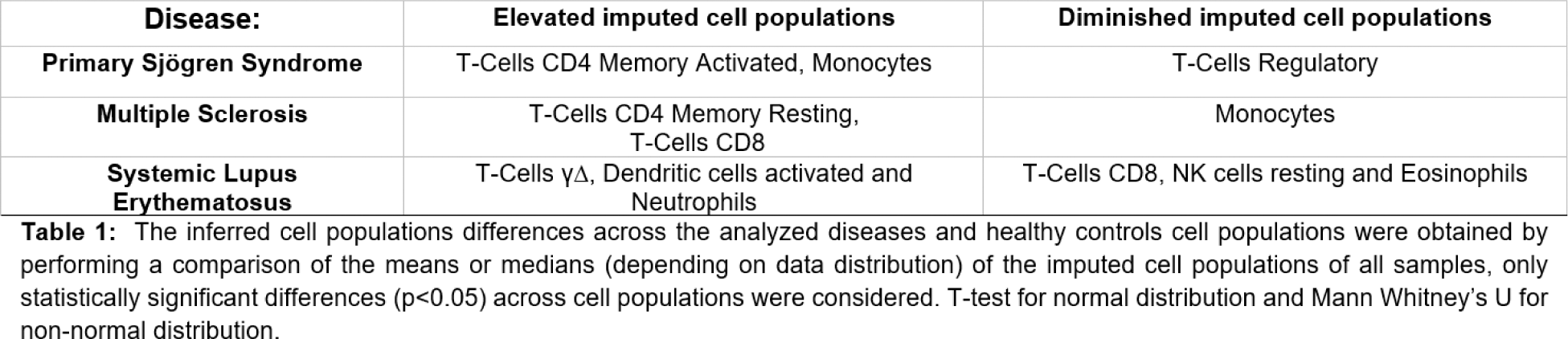
Conclusion: These findings suggest a possible implication of ROS and cellular response to chemical stress in the development or clinical course of the studied autoimmune diseases. The differences observed in the gene expression profiles for each autoimmune disease suggest that the role of the studied pathways may be able to stratify patients of the same disease in smaller groups, possibly characterizing their disease even further. Experimental research is required to explore this hypothesis extensively.
REFERENCES: [1] Tavassolifar, M. J., Vodjgani, M., Salehi, Z., & Izad, M. (2020). The Influence of Reactive Oxygen Species in the Immune System and Pathogenesis of Multiple Sclerosis. Autoimmune diseases, 2020, 5793817.
[2] Di Dalmazi, G., Hirshberg, J., Lyle, D., Freij, J. B., & Caturegli, P. (2016). Reactive oxygen species in organ-specific autoimmunity. Auto- immunity highlights, 7(1), 11.
[3] Ahsan, H., Ali, A., & Ali, R. (2003). Oxygen free radicals and systemic autoimmunity. Clinical and experimental immunology, 131(3), 398–404.
[4] Hänzelmann S, Castelo R, Guinney J (2013). “GSVA: gene set variation analysis for microarray and RNA-Seq data.” BMC Bioinformatics, 14, 7. DOI:10.1186/1471-2105-14-7,
Acknowledgements: We’d like to thank the Mexican National Council of Humanities, Science and Technology (CONAHCYT) for providing the academic grant that made this research possible. Grant ID: PCC-2022-320697.
Disclosure of Interests: None declared.