

Background: Gout is the most common adult inflammatory arthritis. The most important risk factor for the development of gout is elevated serum urate (SU) levels (hyperuricemia). Yet, it remains unclear why only portions of hyperuricemics develop clinical gout.
Objectives: To develop a machine learning-based prediction model for identifying hyperuricemic participants at risk of developing gout.
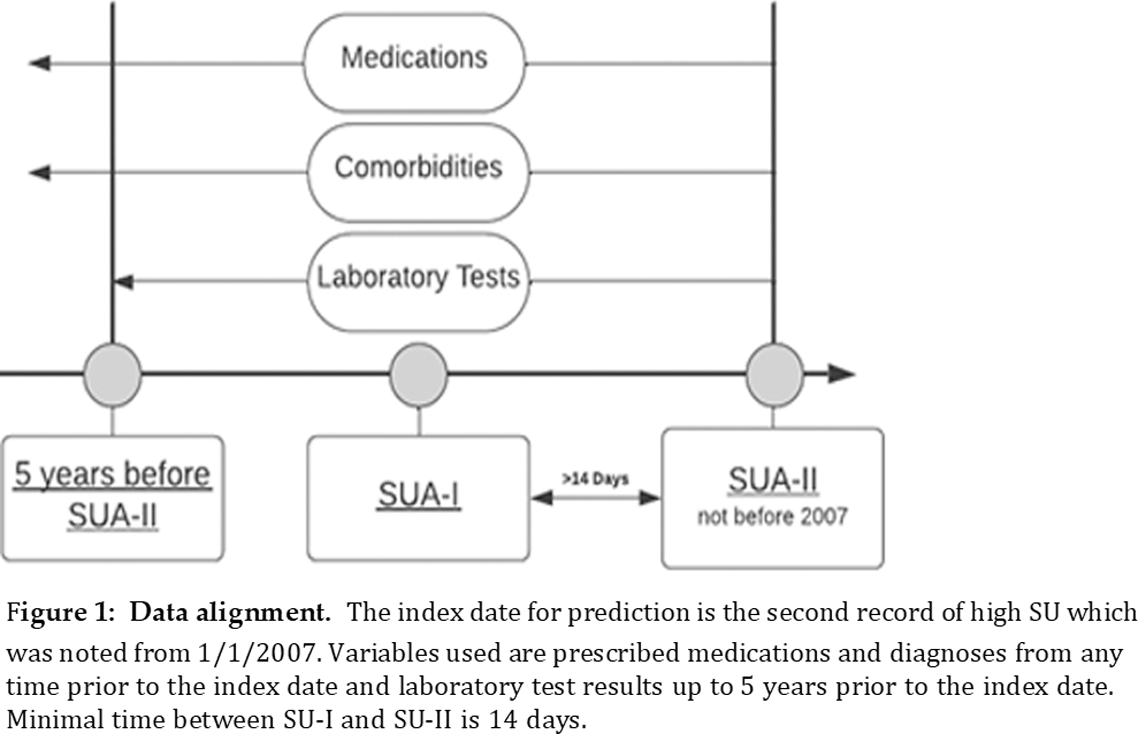
Methods: A retrospective nationwide Israeli cohort study used the Clalit Health Insurance database of approximately 4.8 million people individuals to identify adults with at least two SU measurements exceeding 6.8 mg/dL between January 2007 and December 2022. Patients with a prior gout diagnosis or on gout medications (allopurinol, colchicine, or febuxostat), were excluded. We excluded patients with hyperuricemia not developing gout with less than 3 years of follow-up data to allow for sufficient time to develop gout. Patients’ demographic characteristics, community and hospital diagnoses, routine medication prescriptions, and laboratory results were used to train a risk prediction model. A machine learning model, XGBoost, was developed to predict the risk of gout. Feature selection methods were used to identify relevant variables. The model’s performance was evaluated using the receiver operating characteristic area under the curve (ROC AUC) and precision-recall AUC. The primary outcome was the diagnosis of gout among hyperuricemic patients. We used Shapley Additive Explanations (SHAP) for model interpretation. To have a clinically useful tool, we used the feature selection processes and the SHAP output to examine the performance of the small model with a subset of the top risk factors for gout.
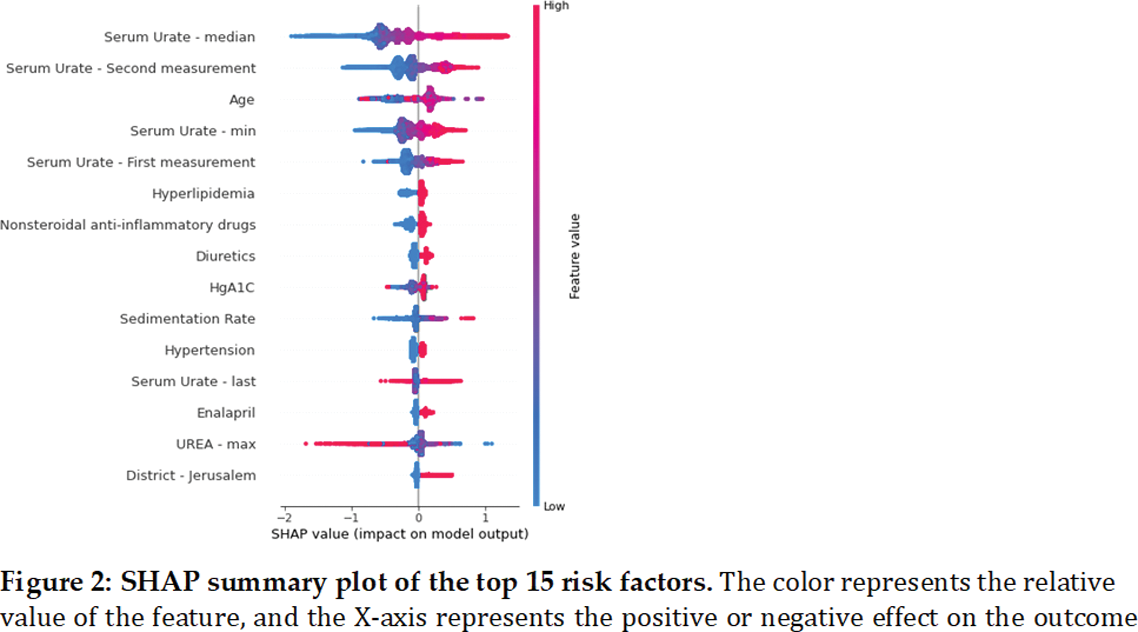
Results: The study cohort comprised 473,124 patients, with 301,385 individuals remaining in the final study population. After applying the exclusion criteria, 15,055 (5%) were diagnosed with gout. The study cohort of hyperuricemic participants included more males (ratio 2.52). This ratio increased among patients with gout (ratio 3.46, p-value< 0.001). The median age was 55 vs. 56 for patients with and without gout (p-value< 0.001). The SU level was higher in gout patients (mean 8.2 mg/dl vs 7.6 mg/dl, p-value < 0.001), and a higher incidence of hyperlipidemia was found in participants with gout (79. 4% vs. 73.6%, p-value < 0.001). The prevalence was higher among patients with SU above 9 mg/dl across all age groups. In total, we had 331 features overall: 7 demographic features, 5 summary values for each of the 21 laboratory test results, 10 binary columns for highly missing laboratory tests, and 2 measurements of SU, 167 medications, and 40 disease binary columns. The XGBoost model had a ROC-AUC of 0.781 (95% CI 0.78-0.784) and precision-recall AUC of 0.208 (95% CI 0.195-0.22). The most significant variables associated with gout diagnosis were SU levels, age, hyperlipidemia, non-steroidal anti-inflammatory drugs and diuretic purchases. A compact model using only these five variables yielded a ROC-AUC of 0.714 (95% CI 0.706-0.723) and a negative predictive value (NPV) of 95%. The calibration curve of the compact model demonstrated a 12.31-fold increase in the percentage of patients with gout between the upper and lower deciles, and the 90th decile had a 13.01 times higher predicted likelihood of developing gout compared to the 10th decile.
Conclusion: To our knowledge, this is the first study that presents a gout prediction model for hyperuricemic patients based on machine learning models. The findings suggest that a our had relatively good performance and high NPV for identifying hyperuricemic participants at risk of developing gout. High NPV means a good prediction of which hyperuricemic will not develop gout and might imply on clinical decisions regarding gout prevention therapy. Our choice of using relatively low SU threshold levels emphasizes the importance of many other variables which are usually available with the use of electronic medical records systems. Altogether, this model should be further validated on different data sets to learn its implications and to further investigate the interactions between variables regarding gout development.
REFERENCES: NIL.
Acknowledgements: We would like to thank our mentor Professor Reuven Mader who passed away during the study period. We would like to thank Mrs. Snait Ayalon from the Emek MC research unit and Shani Shimberg from the rheumatology unit in Emek MC for their great assistance. We would like to thank the” CHS Research Room” team for their support.
Disclosure of Interests: None declared.