

Background: Detailed characterization of the plasma proteome may provide insights into the dynamic molecular changes preceding gout. A previous cross-sectional study of pre-existing gout (n=7 candidate proteins) reported higher CXCL8 levels among prevalent gout than non-gout,[1] but data on pre-diagnostic proteomic biomarkers are sparse.
Objectives: We investigated the relation between large-scale pre-diagnostic proteomics and risk of incident gout in a nationwide prospective cohort, accounting for serum urate levels, and explored biological mechanisms connected to the gout-associated proteins and upstream factors.
Methods: We included 48,898 UK Biobank participants with proteomic measures and no history of gout or urate-lowering therapy use at baseline (2006-10; mean age 56.7 years, 45% males). Proteomic measures (n=2923, via Olink Explore 3072) were quantified from baseline blood samples via Olink’s Proximity Extension Assay technology.[2] 8 proteins with >20% missingness were excluded. Multivariable Cox proportional hazards models were used to assess the association between protein levels and risk of incident gout (from linked primary care and inpatient diagnoses), before and after serum urate adjustment, using a false discovery rate (P FDR ) of <0.05. We performed two-sample Mendelian randomization (MR) to explore potential causal roles of the statistically significant proteins, leveraging genetic association data for loci mapping within 1Mb of the protein-encoding gene controlling expression in cis ,[2] and from the GlobalGout GWAS.[3] We also used Ingenuity Pathway Analysis (IPA) software to explore the canonical pathways and upstream regulators enriched for the gout-associated proteins.
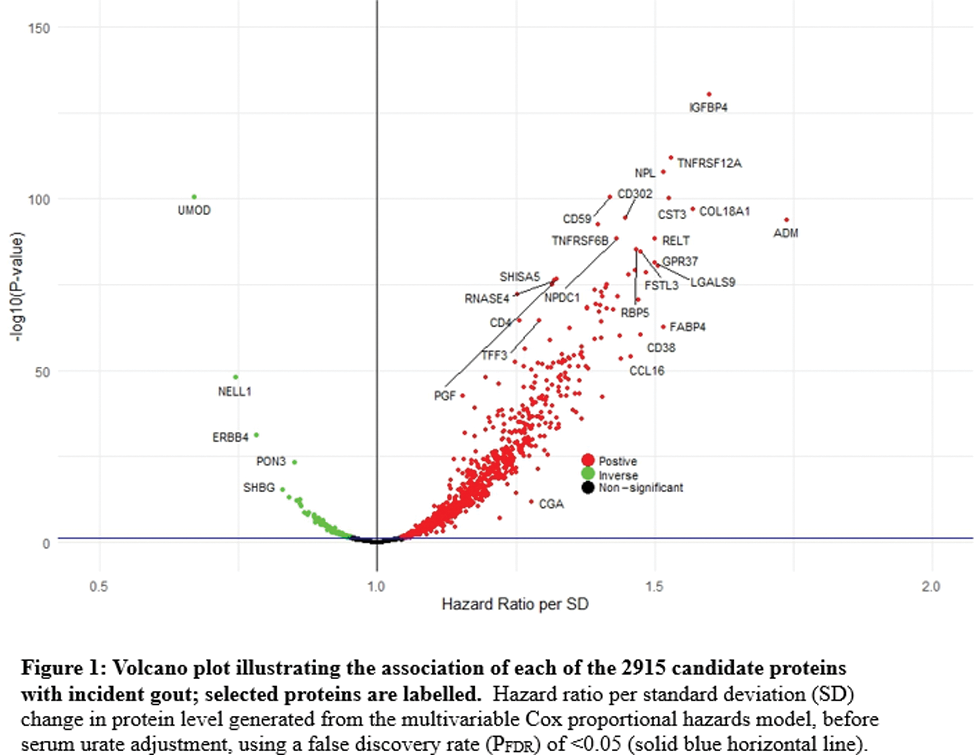
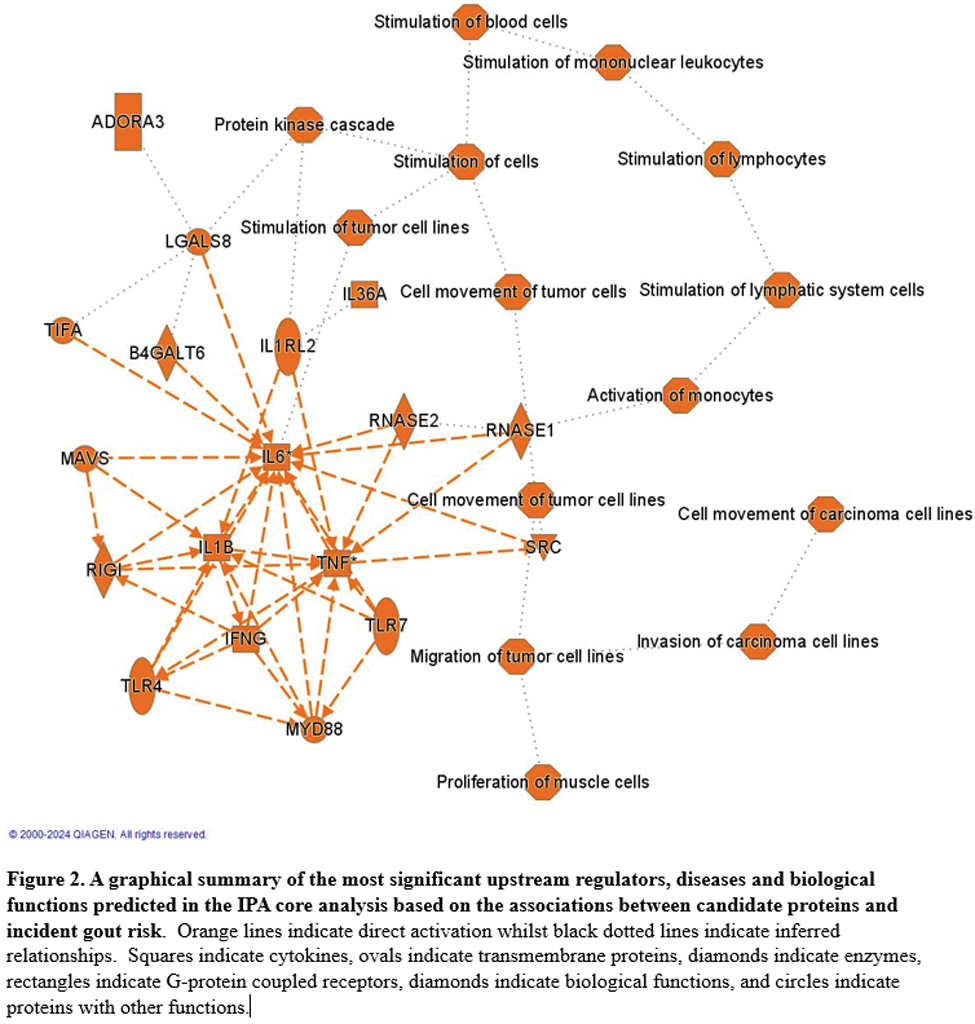
Results: 1095 incident gout cases were documented over mean follow-up of 13.2 years. Levels of 1027 proteins were associated with risk of incident gout before urate adjustment ( Figure 1 ), with the five-strongest associations insulin-like growth factor-binding protein 4 (hazard ratio [HR] 1.60, 95% CI 1.51-1.69 per SD; P FDR 2.5x10 -57 ), TNF receptor superfamily member 12A (1.53, 1.45-1.62; P FDR 2.4x10 -49 ), N-acetylneuraminate lyase (1.52, 1.44-1.60; P FDR 1.5x10 -47 ), uromodulin (0.67, 0.64-0.71; P FDR 2.4x10 -44 ), and CD59 (1.42, 1.35-1.49; P FDR 2.4x10 -44 ). Other key associations were CD38, a proinflammatory NAD-hydrolyzing enzyme expressed in macrophages, CCL16, a substantial monocyte chemoattractant, and ADM, which regulates inflammation bidirectionally, including in the synovium ( Figure 1 ). 41 proteins remained associated with risk of incident gout after serum urate adjustment, including uromodulin, as well as regenerating islet-derived protein 4, N-terminal prohormone of brain natriuretic peptide, sarcoplasmic reticulum histidine-rich calcium-binding protein, MAM domain-containing protein 2, and Spondin-1 (SPON1). MR findings supported causal roles that were directionally consistent for 72 of the 817 measurable proteins associated with gout before urate adjustment, and 3 of 37 measurable proteins after urate adjustment (Secretagogin, SPON1, and TNF receptor superfamily member 6B). Figure 2 is an overview of the most significant upstream regulators and biological functions predicted in the IPA core analysis. Top biological pathways were pathogen induced cytokine storm signalling (P FDR =3.2x10 -48 ), hepatic fibrosis (P FDR =1.3x10 -47 ), and neutrophil degranulation (P FDR =1.3x10 -45 ). Pathways involved in wound healing signalling, regulation of insulin-like growth factor transport and uptake, and granulocyte adhesion and diapedesis were also among the top overrepresented biological pathways. TNF, a cytokine master regulator of inflammatory signaling, was identified as the top upstream regulator of gout-associated proteins. Other upstream regulators included transforming growth factor beta 1 (TGFB1), and IL-1B and IL-4 cytokines.
Conclusion: These prospective findings provide insight on pathophysiological mechanisms leading to gout, including after hyperuricemia, and prioritize protein-gout genetic associations for future research.
REFERENCES: [1] PMID 26248007.
[2] PMID 37794186.
[3] 10.1101/2022.11.26.22281768.
Acknowledgements: NIL.
Disclosure of Interests: Natalie McCormick: None declared, Amit Joshi Regeneron Pharmaceuticals, Regeneron Pharmaceuticals, Robert Terkeltaub Allena, LG Chem, Fortress/Urica, Selecta Biosciences, Horizon Therapeutics, Atom Bioscience, Acquist Therapeutics, Generate Biomedicines, Astra-Zeneca, and Synlogic (current or recent), Astra-Zeneca (previously), Tony Merriman: None declared, Chio Yokose: None declared, Hyon hoi Ani, LG, Horizon, Shanton, Protalix, Horizon.