

Background: Osteoarthritis (OA) is a degenerative joint disease affecting multiple tissues in the joint including the synovium. The synovium is a connective tissue ensuring joint lubrication however, in OA, the synovium undergoes substantial changes including inflammation, hyperplasia and cellular proliferation. This study sought to identify key cellular components involved in OA synovial pathology utilizing single-nuclei RNA sequencing (snRNA).
Objectives: 1. Identify cell types and distinct subtypes in the synovium of early (KL1) and late (KL3/4) stage radiographic knee OA and their associated transcriptomic signatures using snRNAseq.
2. Determine the presence of identified human OA synovium cell subtypes in the synovium of destabilization of the medial meniscus (DMM)-OA mice using snRNAseq of the murine synovium.
3. Investigate the contribution of key transcriptional regulators in ECM/fibrosis regulatory mechanisms in vitro and in vivo.
Methods: SnRNA sequencing was performed on human OA knee synovial tissues from early- (KL1; n=5) and late- (KL3/4; n=4) stage radiographic knee OA patients and mouse synovia from naïve as control (n=3), 2 weeks (n=4) and 10 weeks (n=3) post destabilization of the medial meniscus (DMM) surgery. Bulk RNA sequencing was performed on human OA synovium from patients with KL1 (n=6) and KL3/4 (n=8) radiographic knee OA. Flow cytometry was performed on cryopreserved synovium from healthy donors (n=5), early (n=10) and late (n=14). Bioinformatics analysis was performed to discern cell types and subtypes using canonical markers, differentially expressed genes (DEG), key pathways, and putative transcriptional regulators implicated in disease progression.
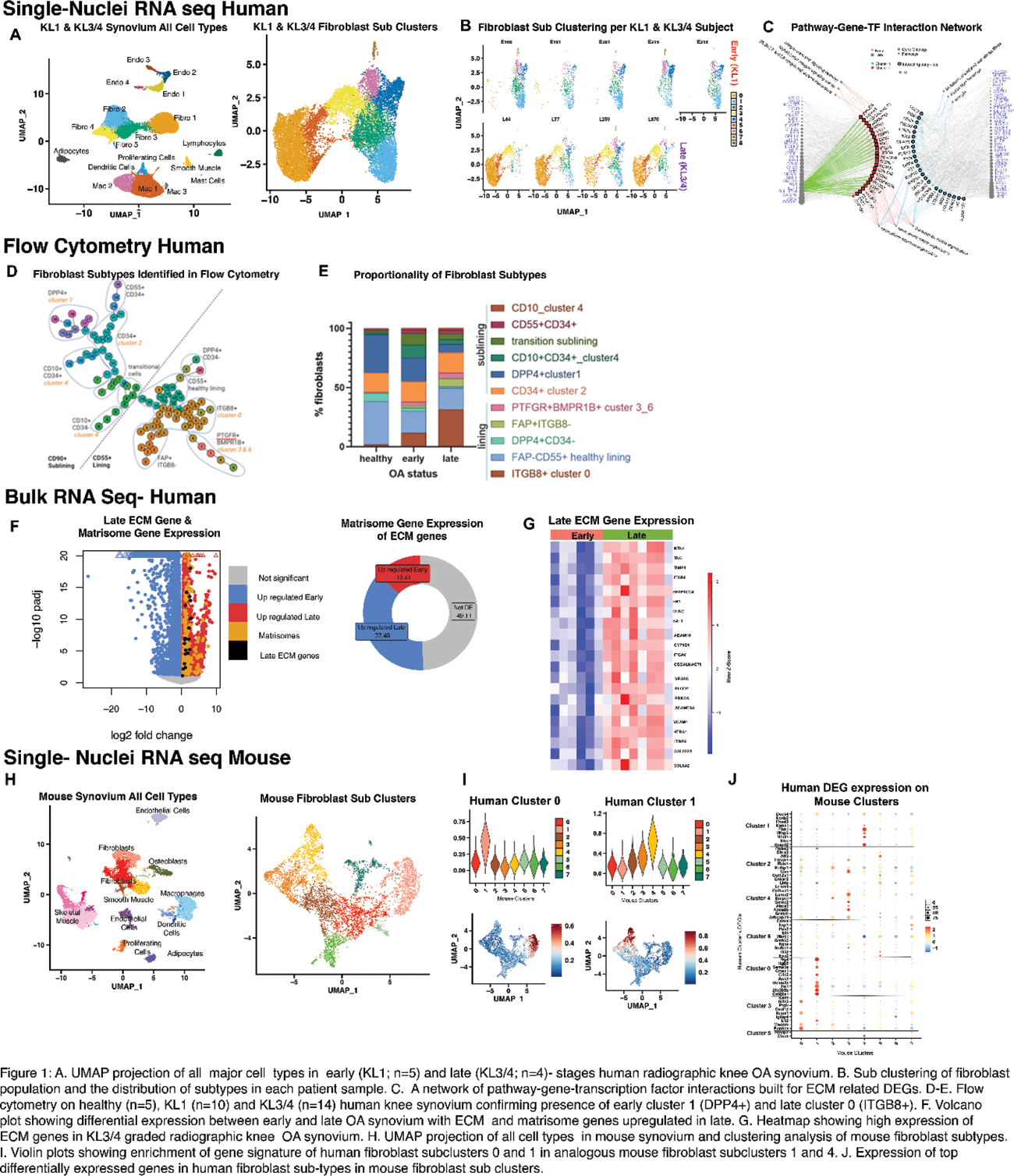
Results: SnRNA sequencing of 25285 nuclei from human knee synovial tissue identified fibroblasts to be the predominant cell type, comprising at least 50% of the cellular composition from KL1 and KL3/4 graded radiographic knee OA synovium. Clustering analysis of fibroblasts resolved nine fibroblast subtypes. An endophenotypic shift in fibroblast subsets was noted with disease stage. Synovia from early-stage OA contributed a higher proportion of nuclei to subclusters 1, 2, 4 and 6 while late-stage synovium predominantly contributed nuclei to subclusters 0, 3 and 5 ( FIGURE 1A&B ). Flow cytometry confirmed a graded expansion of ITGB8+ (cluster 0) lining fibroblasts and a contraction of DPP4+ (cluster 1) sub-lining fibroblasts with OA progression ( FIGURE 1D&E ). A pathway analysis was performed on human fibroblast subclusters highlighting that cluster 0 and 1 are primarily involved in pathways related to the extracellular matrix (ECM)( FIGURE 1C ). To confirm these findings, we conducted bulk RNA sequencing on KL1 and KL3/4 graded human synovium, confirming that genes upregulated in late-stage synovial samples show enrichment for matrisome-related genes ( FIGURE 1F&G ). A regulatory transcription factor (TF) prediction analysis was performed on ECM genes from the DEG lists of human clusters 0 and 1 using Catrin. This identified three putative upstream TFs, namely PGR, ELF1 and BHLHE40, specifically associated with late-stage cluster 0. To profile the temporal dynamics of key cell subtypes the DMM mouse model characterized the presence of cell subsets and their transcriptomic profiles analogous to those found in human synovia. The snRNA sequencing of 19304 nuclei from DMM/naïve mouse synovium revealed comparable fibroblast subtypes to those identified in humans. Specifically, the largest late-stage human fibroblast subcluster (cluster 0) was most similar to mouse cluster 1, while the prevalent early-stage human fibroblast subcluster (cluster 1) most resembled mouse fibroblast cluster 4 ( F1GURE 1H-J ).
Conclusion: We have identified an endophenotypic shift in fibroblast subsets from early to late stages of radiographic knee OA that may play a crucial role in synovial ECM regulation and fibrosis during OA. Subsequent investigations will elucidate if the identified TF’s control ECM regulation and fibrosis through in vitro and in vivo knock out models.
REFERENCES: NIL.
Acknowledgements: This work is funded by the Canadian Institute of Health Research Operating Grant, Tony and Shari Fell Platinum Chair in Arthritis Research and Canada Research Chairs Program.
Disclosure of Interests: None declared.