

Background: VEXAS syndrome is an adult-onset, X-linked, life-threatening, autoinflammatory disease with predominant hematological involvement caused by somatic mutation in UBA1 gene whose pathophysiology is still unknown.
Objectives: To achieve a molecular and phenotypic characterization of hematopoiesis of VEXAS patients and to develop cellular and humanized mouse models by gene editing.
Methods: Six VEXAS patients (p.Met41>Thr; p.Met41>Val; p.Met41>Leu; c.118-1 G>C) were recruited from our Unit. Variant allele frequency (VAF) of UBA1 mutant cells was quantified by targeted sequencing in isolated hematopoietic lineages and hematopoietic stem/progenitor cells (HSPCs). Multiparametric immunophenotypic analysis and single cell RNA seq were performed on peripheral blood and bone marrow (BM), focusing on HSPCs. Circulating monocytes were analyzed by whole RNA-seq and metabolome analysis. Healthy age and sex-matched controls were included. To introduce UBA1 mutations and develop VEXAS models, cutting-edge gene editing technologies were adopted in healthy human HSPCs.
Results: Targeted sequencing in VEXAS patients showed >0.8 VAF in HSPCs. Conversely, VAF largely differed across mature cells, averaging 0.81 in neutrophils, 0.64 in monocytes, 0.42 in NK, 0.07 in T cells, and 0.09 in B cells, supporting a myeloid skewing of mutant HSPCs. Multiparametric immunophenotypic analyses showed unbalanced composition of HSPCs in the BM, with 2-to-3-fold reduction of primitive stem cells, multipotent and lymphoid progenitors, and 2-fold increase of myeloid progenitors, compared to matched healthy individuals. HSPCs, myeloid-biased HSPCs and immature myeloid cells were increased by 3-to-4 fold in the circulation (p<0.03). Gene expression analysis of circulating monocytes displayed upregulation of inflammatory pathways and metabolic rewiring (Figure. 1, panels A-B). Metabolomic analyses confirmed hyperactivation of the glycolytic pathway and specific lipid metabolism (Figure. 1, C-D). Single-cell RNA-seq of bone marrow mononuclear cells identified a subpopulation of CD34+ cell specific of VEXAS patients and revealed upregulated stress response and immune activation pathways across VEXAS cell clusters compared with healthy controls. Models of VEXAS generated by gene editing recapitulated patients’ hematopoiesis and pathophysiology in vitro and in vivo . UBA1 mutations were installed at VAF>0.9 in HSPCs and generated a myeloid bias in vitro . Transplantation of edited HSPCs in immunodeficient mice resulted in a 100-fold reduction in circulating B cells, while NK and myeloid compartments were preserved. Human BM HSPCs were 5-fold lower than control mice, largely myeloid-biased and presented abnormal vacuolar morphology. Concordantly, VAF was >0.8 in myeloid cells and HSPCs and <0.3 in B cells. Figure 1. Panel A. Gene expression analysis of peripheral monocytes in VEXAS patients compared to controls. Panel B. Gene-enrichmed pathway analysis in VEXAS patients compared to controls. Panel C. Metabolic analysis in VEXAS patients compared to controls. Panel D. Metabolic-enriched pathway analysis in VEXAS patients compared to controls.
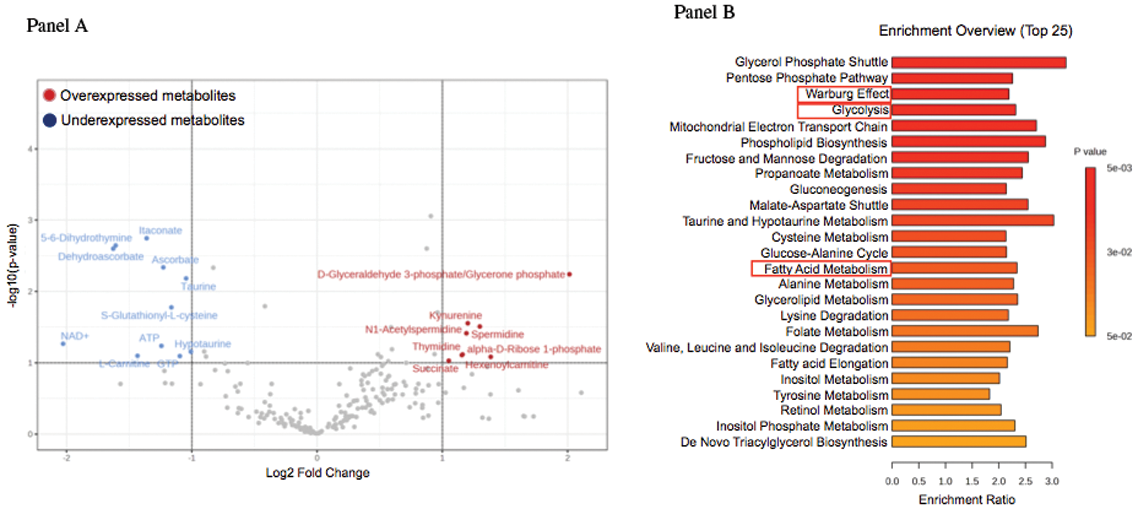
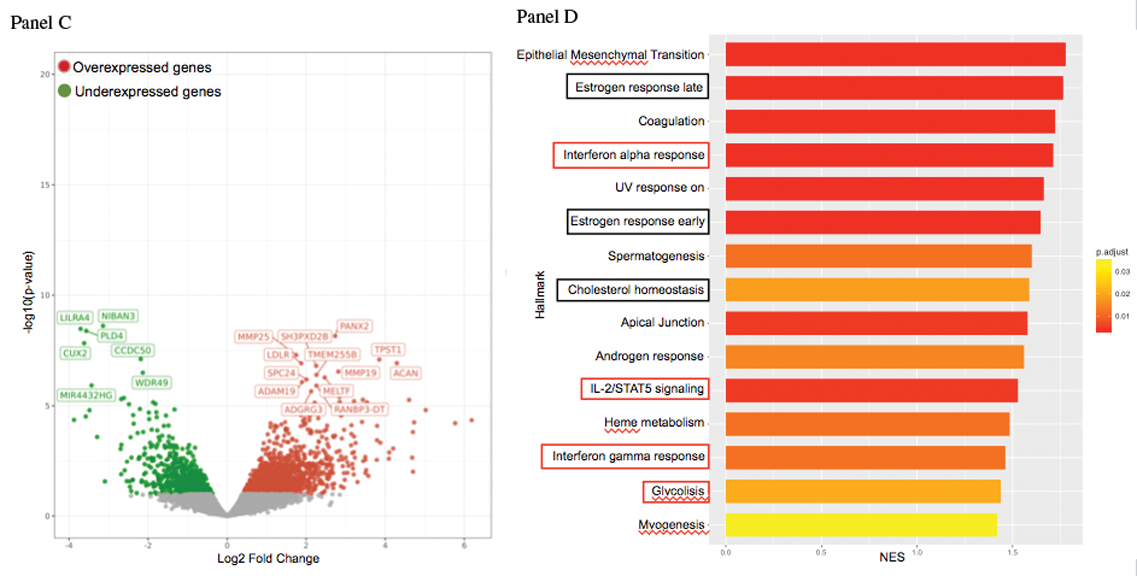
Figure 1.
Conclusion: Mutations in UBA1 drive expansion of HSPCs and enhance myelopoiesis-guided accumulation of myeloid precursors. Mutant lymphoid cells are negatively selected and their myeloid counterpart in peripheral blood displays upregulation of transcriptomic signatures and metabolic pathways indicative of inflammatory activation. Gene editing-based models hold promise to enable preclinical testing and validation of novel therapeutics to treat VEXAS syndrome.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: Corrado Campochiaro: None declared, Raffaella Molteni: None declared, Guido Pacini: None declared, Martina Fiumara: None declared, Alessandro Tomelleri: None declared, Elisa Diral: None declared, Davide Stefanoni: None declared, Angelica Varesi: None declared, Alessandra Weber: None declared, Roberta Alfieri: None declared, Luisa Albano: None declared, Maddalena Panigada: None declared, Eleonora Cantoni: None declared, Daniele Canarutto: None declared, Luca Bassoricci: None declared, Pamela Quaranta: None declared, Angelo D’Alessandro: None declared, Marco Matucci-Cerinic: None declared, Raffaella Di Micco: None declared, Alessandro Aiuti: None declared, Fabio Ciceri: None declared, Ivan Merelli: None declared, Lorenzo Dagna: None declared, Serena Scala: None declared, Simone Cenci: None declared, Luigi Naldini: None declared, Samuele Ferrari: None declared, Giulio Cavalli Novartis, Novartis.