

Background: EGPA is a rare systemic autoimmune disorder, included within the ANCA-associated vasculitis. It is characterized by a diverse clinical profile, together with an unsatisfactory response to treatment leading to frequent relapses, with chronic asthma being the most recurrent type [1]. Its pathogenesis remains unclear, being supported by experimental studies with limited evidence. However, they all indicate that EGPA pathogenesis is mainly driven by Th2 lymphocytes (CD4 + T cells), showing oligoclonal expansion in active patients, along with a significant increase in serum levels of chemokines involved in Th2 recruitment and activation [2]. Moreover, some genetic variants associated to Th2 activation have been identified in these patients [3]. In fact, Th2 lymphocytes are known to be the main responsible cell type for the activation of eosinophils, the core effector cells of the disease [4].
Objectives: To investigate transcriptomic changes of Th2 lymphocytes in patients with EGPA in order to characterize the molecular basis of the heterogeneity of this disease, and to identify pathogenic pathways that could provide clinically useful biomarkers.
Methods: CD4 + T lymphocytes were isolated from EGPA patients in remission (BVAS = 0, prednisone dose <7.5 mg/day) (n=23) and compared to healthy controls (n=14) and non-EGPA asthmatic patients (n=8). Affymetrix Clariom TM S human microarray was used to analyze whole transcriptome signature. Unsupervised hierarchical clustering and principal component analysis (PCA) were conducted to identify clusters among patients with similar features. Analysis of transcriptomic changes (GSEA) (p < 0.001, FC >1.5) among the different groups, and GO enrichment analysis (ShinyGo, KEGG) of the differentially expressed genes (DEGs) were performed. The most interesting DEGs were validated by qPCR. Two-sample permutation T-test was used for statistical analyses through BRB-ARRAY Tools.
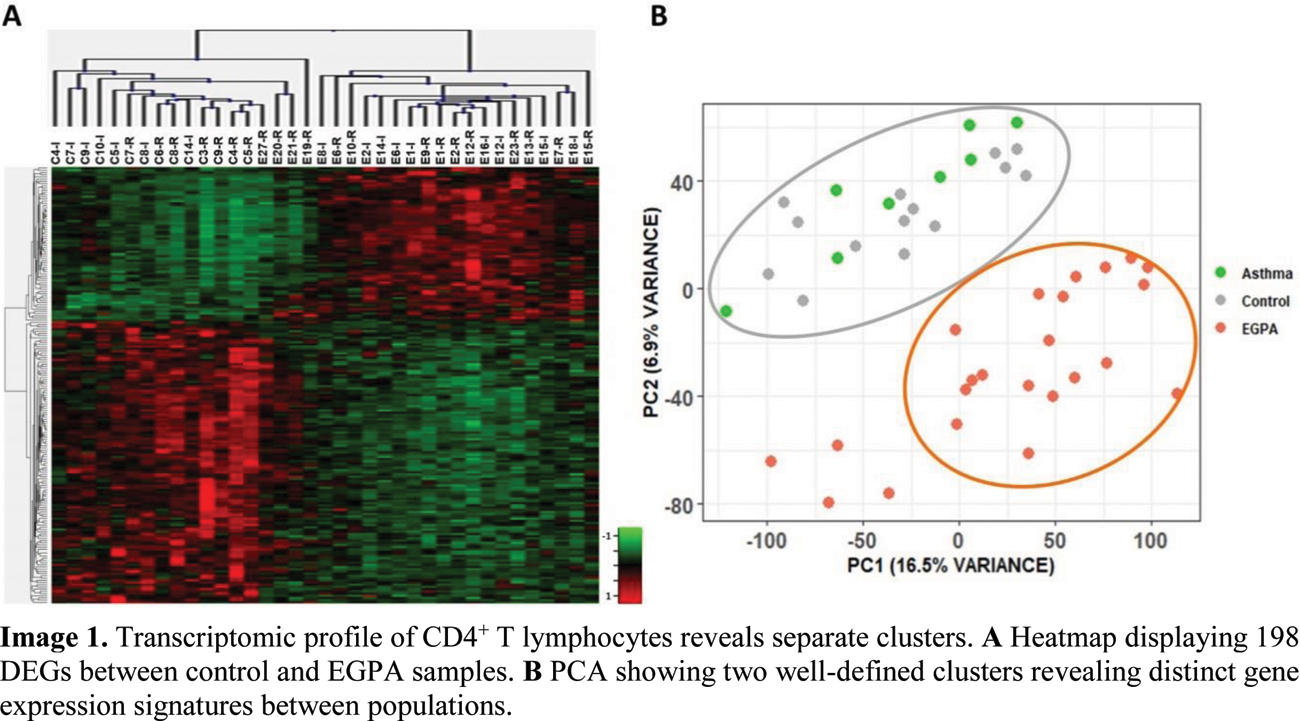
Results: 198 DEGs were found between EGPA and control samples, defining two distinct clusters as evidence in the heatmap (Figure 1A). PCA of all samples included in the analysis further confirmed these two groups (Figure 1B). Pathway enrichment analysis showed significant differences between these two populations regarding Th1, Th2 and Th17 cell differentiation, IL2/STAT5 pathway and interferon α (IFN α) and IFNγ response. qPCR specifically validated IL2/STAT5 pathway through various relevant genes, such as CTLA4, CD81, and LTB, suggesting a potential role in the pathogenesis of EGPA.
Transcriptomic profile of CD4 + T lymphocytes reveals separate clusters. A Heatmap displaying 198 DEGs between control and EGPA samples. B PCA showing two well-defined clusters revealing distinct gene expression signatures between populations.
Conclusion: Our study provides transcriptomic evidence that CD4 + T cell expression profile is modified in EGPA patients. GSEA and DEGs show a potential role of different pathways involved in CD4 + T lymphocyte differentiation and activation, potentially mediated by IL2/STAT5 pathway.
REFERENCES: [1] Wechsler ME et al . J Allergy Clin Immunol. 2023;151(6):1415-1428.
[2] Isozaki T et al . J Clin Med. 2020;9(12):3890.
[3] Lyons PA et al . Nat Commun. 2019; 10(1):5120.
[4] Jakiela B et al . Rheumatology (Oxford). 2012;51(10):1887-93.
Acknowledgements: This study was supported by PI18/00461 ISCIII-Subdireccion General de Evaluacion, Fondo Europeo de Desarrollo Regional (FEDER) “Otra manera de hacer Europa”.
Disclosure of Interests: Roberto Ríos-Garcés: None declared, Núria Farran: None declared, Salvador Naranjo-Suárez: None declared, Roser Alba-Rovira: None declared, Sergio Prieto-González: None declared, Itziar Tavera-Bahillo: None declared, Ebymar Arismendi: None declared, Roser Solans-Laqué: None declared, Marc Corbera-Bellalta: None declared, Farah Kamberovic: None declared, Nina Visocnik: None declared, Maria C. Cid GSK, AstraZeneca, AbbVie and CSL-Vifor, GSK, AstraZeneca, AbbVie and CSL-Vifor, Kiniksa Pharmaceuticals Corp., Georgina Espígol-Frigolé Vifor and GSK, Vifor, GSK and Janssen, GSK.