

Background: The exact origin and precursor differentiation route of tissue macrophages remains controversial. Deep characterisation of myeloid cell subsets by single cell RNA sequencing (scRNA-seq) across healthy and inflamed tissues in rheumatoid arthritis (RA) has led to the identification of new pathogenic cell states and subsets in five recent large-scale studies ([1-5] including from the Accelerating Medicines Partnership (AMP) RA Consortium [4,5]). However, subset overlap across studies and compartments (blood versus synovial tissue) has not yet been systematically investigated.
Objectives: To map monocyte subsets and states across studies and compartments to identify blood monocyte precursors of inflammatory synovial macrophage subsets observed in RA.
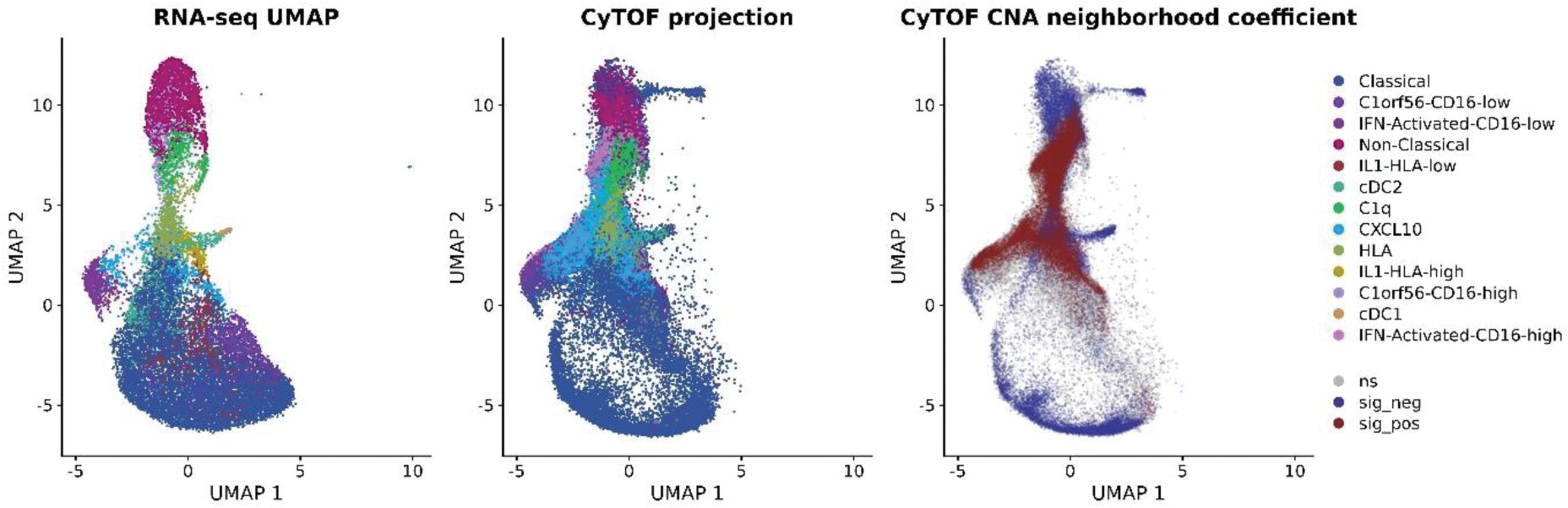
Methods: First, peripheral blood mononuclear cells (PBMCs) from healthy volunteers and RA patients with clinically well-controlled disease (quiescent PBMCs) were enriched for monocytes by negative selection and subjected to scRNA-seq (10X Genomics). Clustering of 20,746 cells was performed in Seurat (generation of a Uniform Manifold Approximation and Projection (UMAP) template). Second, published myeloid cell subsets (comprising a total number of 110,351 myeloid cells from 5 scRNA-seq studies [1-5]) were mapped onto this template based on the similarity of their expression scores. Hierarchical clustering was applied to merge similar clusters to create a consensus map. Third, random forests were used as a novel method of merging over-clustered data to identify novel myeloid cell states and generate a final taxonomy of monocyte states in human healthy blood ( Figure 1 , left panel, with CD14 high classical monocytes in dark blue at the bottom, CD16 + non-classical monocytes in magenta at the top, intermediate monocytes in the middle). Finally, to provide experimental validation at the protein level, PBMCs from 19 RA patients with uncontrolled inflammation (DAS > 5.1 and treatment failure with conventional Disease Modifying Anti-Rheumatic Drugs) were deeply immunophenotyped with a 23-marker myeloid panel by mass cytometry (CyTOF). Inflammatory cell states with increased abundance in RA were identified with Co-varying Neighborhood Analysis (CNA) [6]. The CyTOF dataset was mapped back onto the scRNA-seq template ( Figure 1 middle panel) using bridge integration implemented in Seurat v5, using the COvid-19 Multi-omics Blood ATlas (COMBAT) Consortium CITE-seq dataset [7] as a bridge.
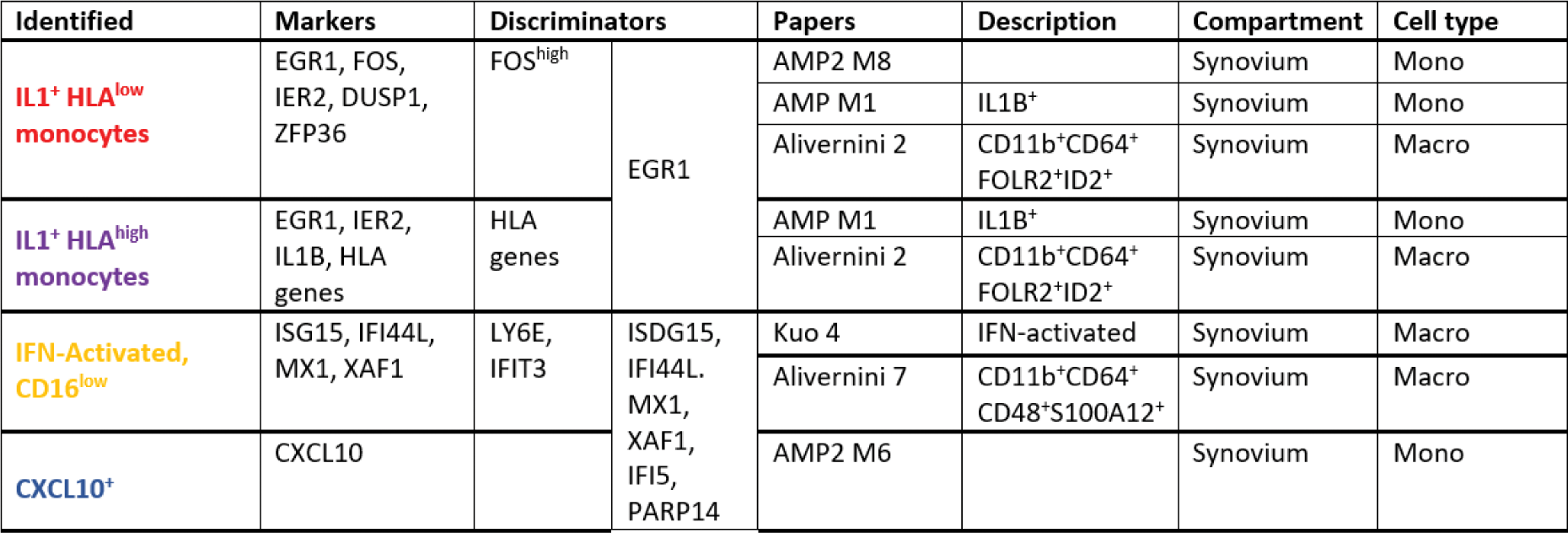
Results: We generated an exhaustive reference atlas comprising a total of 11 monocyte states across anatomical compartments relevant for RA ( Figure 1 , left panel). For example, we show that the CD11b + CD64 + FOLR2 + ID2 + cluster in [3], the IL1B + cluster in [4] and cluster M8 in [5] represent the same inflammatory synovial macrophage subset (first of the 4 examples featured in Table 1 ) and are transcriptionally similar to an IL1B + monocyte subset present in quiescent peripheral blood. Next, we show that 4 quiescent monocyte states present in the peripheral blood of both patients and healthy individuals (IL1B + , CXCL10 + , C1Q + , and IFN-activated, all originating from CD14 + CD16 + intermediate monocytes ( Figure 1, left panel) expand in the blood of patients with uncontrolled RA ( Figure 1 , middle panel). The statistically significant cell clusters (FDR<0.05) are colour-coded in dark red on Figure 1 , right panel.
Conclusion: We define a new monocyte cell taxonomy relevant for RA comprising a total number of 11 continuous cell states dynamically transitioning into each other across anatomical compartments. We show that 4 quiescent peripheral blood intermediate monocyte states, sharing a transcriptional signature with inflammatory synovial macrophages, expand in uncontrolled RA and therefore likely represent blood precursors of pathogenic tissue macrophages.
REFERENCES: [1] Villani AC, Science, 2017.
[2] Kuo D, Sci Transl Med, 2019.
[3] Alivernini S, Nat Med, 2020.
[4] Zhang F, Nat Immunol, 2019.
[5] Zhang F, Nature, 2023.
[6] Reshef YA, Nat Biotechnol, 2022.
[7] COMBAT Consortium, Cell, 2022.
Table 1.
Acknowledgements: BRAGGSS Collaborators.
Disclosure of Interests: None declared.