

Background: An accurate prediction of the response of patients with arthralgia at risk for rheumatoid arthritis (RA) to preventive treatment could impact treatment decision making and reduce the risk of overtreatment. Assuming that MRI scans of the wrist, hand and foot contain sufficient information to predict response to treatment, we hypothesized that employing artificial intelligence methods, such as deep learning, in analyzing these images could predict early treatment response in patients with clinically suspect arthralgia.
Objectives: Therefore, we aimed to develop a deep learning model and assess its ability to predict treatment response in patients with clinically suspect arthralgia, based on MRIs of the wrist, MCP and MTP joints at baseline from the TREAT-EARLIER trial [1].
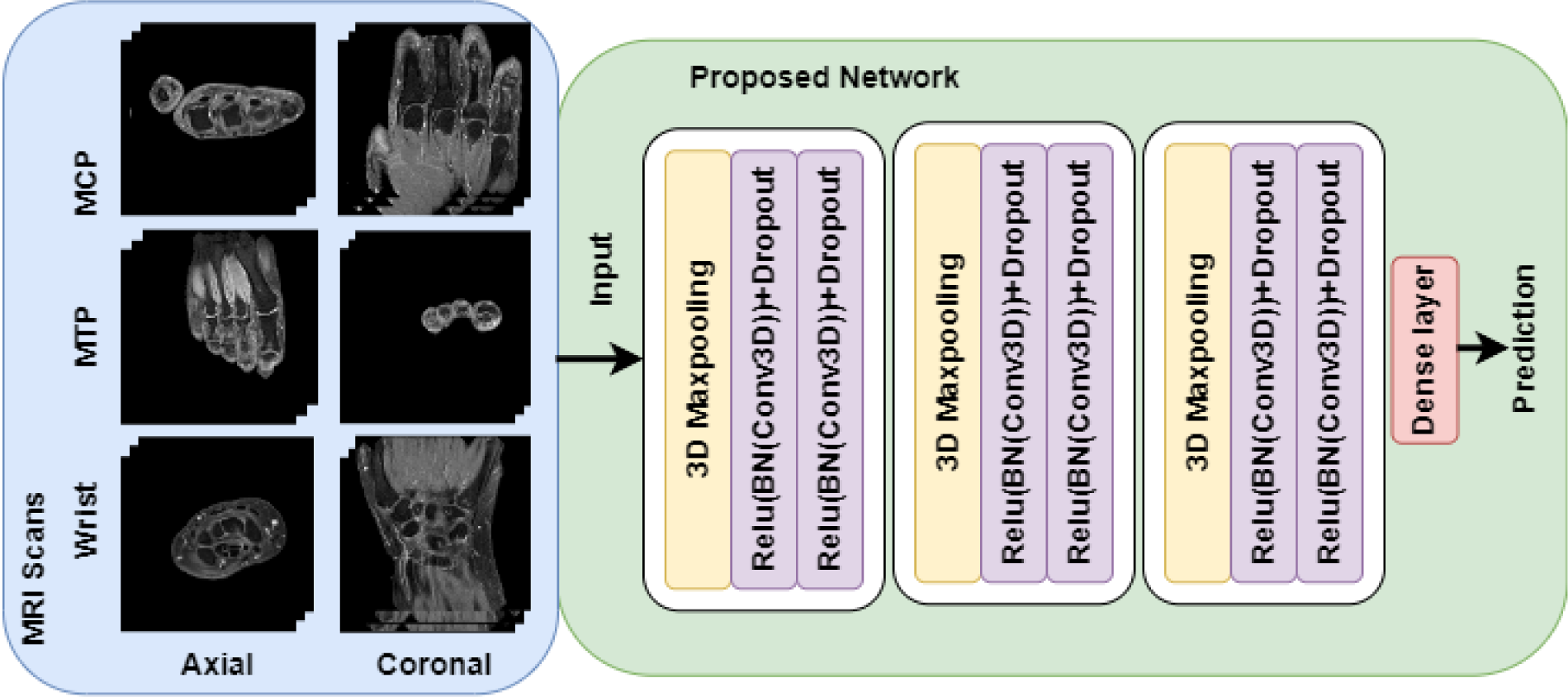
Methods: MRI scans (with contrast-enhanced T1-weighted TSE fat-suppressed sequences) were collected from 236 patients with clinically suspect arthralgia at baseline, from the TREAT-EARLIER trial that determined the response to treatment by a single intramuscular methylprednisolone injection followed by methotrexate during one year[1]. Patients were assigned to a placebo (117) or treatment (119) group. Treatment response was defined as a decrease in subclinical joint inflammation scores (according to RAMRIS) greater than the 5th percentile of the decrease between baseline and 12-months in the placebo group. By using this threshold, we corrected for possible changes in subclinical inflammation scores due to spontaneous variations or regression to the mean. From 103 patients in the treatment group, both axial and coronal MRI scans of the wrist, MCP and MTP joints were available. These scans were first resized to 128 × 128 × 14 pixels and the background outside the hand or foot was removed. Subsequently, a 3D convolutional neural network (Figure 1) was trained with these baseline MRI scans to predict treatment response after one year. To evaluate the proposed model, full 10-fold cross-validation was performed five times with different random splits, and the average and standard deviation of the area under the receiver operating characteristic curve (AUC) was determined. From the average confusion matrix, the test characteristics were calculated.
Results: In the placebo group, the 5th percentile of the change in subclinical inflammation scores was -1 for each of the categories synovitis, tenosynovitis and bone marrow edema. A decrease greater than -3 was therefore considered a reference for a significant treatment response. This threshold is in accordance with the smallest detectable change in our data. The five 10-fold cross-validations showed that the proposed model could predict treatment response with an 80% ± 0.021 AUC based on the baseline (axial and coronal) MRI scans of the wrist, MCP and MTP joints. The positive predictive value was 0.84, the negative predictive value was 0.78, sensitivity was 0.78, and specificity was 0.84.
Conclusion: The results suggest that the proposed deep learning model can predict treatment response in patients with arthralgia at risk for RA development from baseline MRI scans. Future work will focus on whether adding clinical data to the deep learning would further improve the prediction. Current data are promising in suggesting that deep learning is helpful in arriving at personalized interventions and reducing the risk of overtreatment in patients with arthralgia that are suspected to develop RA.
REFERENCES: [1] Krijbolder, DI, et al. “Intervention with methotrexate in patients with arthralgia at risk of rheumatoid arthritis to reduce the development of persistent arthritis and its disease burden (TREAT EARLIER): a randomised, double-blind, placebo-controlled, proof-of-concept trial.” The Lancet 400.10348 (2022): 283-294.
Overview of the proposed model.
Acknowledgements: This AI-project has been funded by the Dutch Research Council (NWO) Applied and Engineering Sciences (project number 17970). The TREAT-EARLIER trial has been funded by an NWO-ZonMW grant (project number 95104004). The Dutch Arthritis Society contributed financially to both grants.
Disclosure of Interests: Tahereh Hassanzadeh Grant/research support from: Bristol-Myers Squibb and Pfizer contributed to this project, through a grant from the Dutch Research Council (NWO), Applied and Engineering Sciences., Yanli Li: None declared, Denis Shamonin Grant/research support from: Bristol-Myers Squibb and Pfizer contributed to this project, through a grant from the Dutch Research Council (NWO), Applied and Engineering Sciences., Dennis A. Ton: None declared, Monique Reijnierse Grant/research support from: Bristol-Myers Squibb and Pfizer contributed to this project, through a grant from the Dutch Research Council (NWO), Applied and Engineering Sciences., Annette H.M. van der Helm – van Mil Grant/research support from: Bristol-Myers Squibb and Pfizer contributed to this project, through a grant from the Dutch Research Council (NWO), Applied and Engineering Sciences., Berend Stoel Grant/research support from: Bristol-Myers Squibb and Pfizer contributed to this project, through a grant from the Dutch Research Council (NWO), Applied and Engineering Sciences.