

Background: Sjögren’s disease (SjD) presents an unmet medical challenge as there is currently no cure. Despite advances in understanding the immunopathogenesis of SjD, there is still a pressing need to identify novel biomarkers and therapeutic targets, for better patient stratification and personalized treatment.
Objectives: To create a fully-detailed molecular interaction map (MIM) including all the signalling and molecular pathways implicated in SjD pathogenesis. To create a large-scale mechanistic model to enable in silico simulations of perturbations including drug interventions, and the generation of hypothesis-driven predictions.
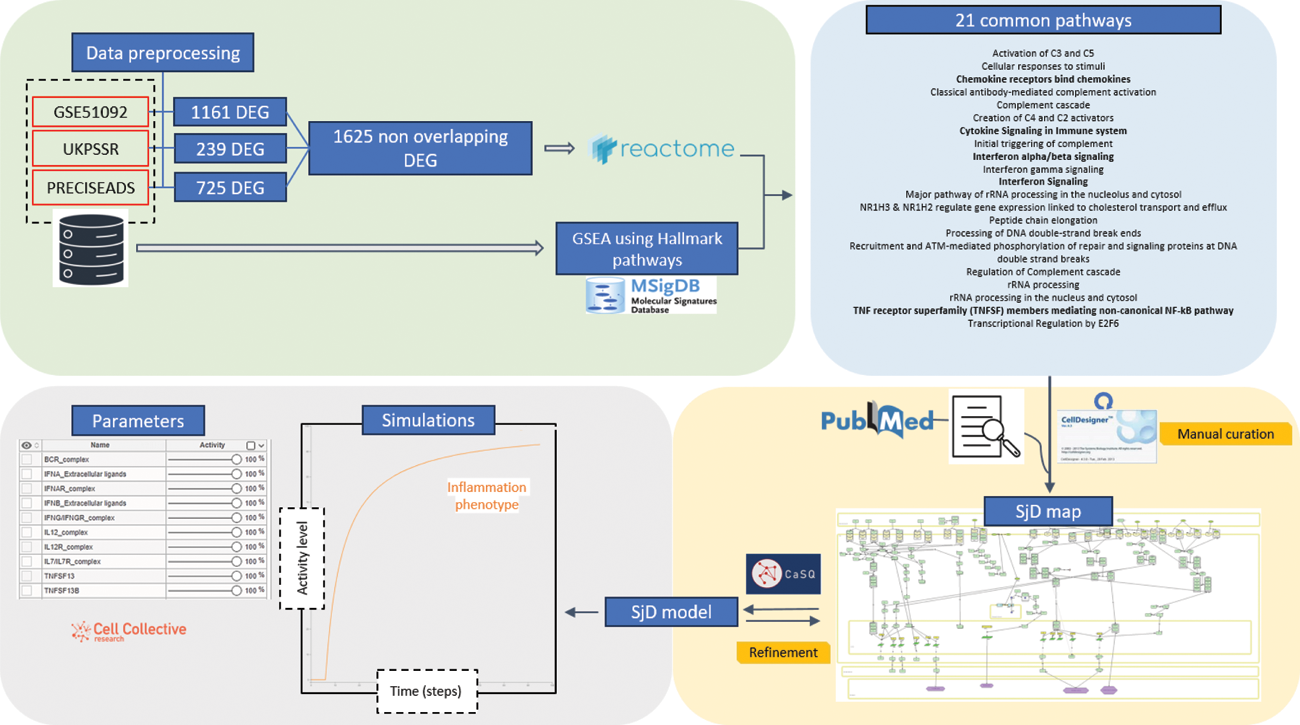
Methods: Differential expression analysis was performed on blood samples from SjD patients vs controls on 3 datasets: the publicly available GSE51092 and the accessible via the NECESSITY consortium UKPSSR and PRECISEADS datasets. GSE51092 contains transcriptomic data of 190 SjD patients and 32 controls, UKPSSR of 151 SjD patients and 29 controls, and PRECISESADS, RNASeq data for 304 SjD patients and 341 controls. Pathway enrichment analysis was subsequently performed using GSEA and the Reactome pathway database[1,2]. Additional literature-based evidence was used to develop a molecular interaction map, combining the results of the previous analytical steps. The SjD specific map was then converted into a Boolean model using the CaSQ tool[3]. Logic rules based on Boolean algebra are used to describe every interaction between the molecular entities.
Results: Our analysis unveiled a set of differentially expressed genes (DEG) and related pathways associated with immune dysregulation and inflammatory responses in SjD. We obtained a total of 1625 DEG, 725 DEG from PRECISESADS, 1161 DEG from GSE51092 and 239 DEG from UKPSSR, with 25 DEG common for all three datasets. Nine common DEG were associated with Interferon signalling. Twenty-one pathways were identified with both GSEA and Reactome-based analysis. The building of the SjD MIM was performed based on literature search and the 21 identified pathways, from the data analysis. The MIM includes so far 16 pathways (5 present in the identified pathways and 11 literature-mined), comprising 187 species (genes, RNAs proteins) and 132 reactions. The SjD-specific Boolean model obtained after conversion of the SjD MIM, represents a more compact and fully executable version of the SjD molecular network, containing 111 nodes and 130 edges. In Sjögren-specific conditions the model is able to reproduce the activation of main inflammation pathways and predict the inflammatory response.
Conclusion: We have built the first SjD-specific MIM integrating omic data analyses and information from literature-based evidence and pathway enrichment analysis. The current SjD map contains hallmark disease pathways and is constantly being enriched with data-driven highlighted pathways. The preliminary computational model based on the SjD map is able to reproduce inflammatory response scenarios, however further training is needed to improve performance and robustness.
REFERENCES: [1] Fabregat, A. et al. The Reactome pathway Knowledgebase. Nucleic Acids Research 44, D481–D487 (2016).
[2] Subramanian, A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 102, 15545–15550 (2005).
[3] Aghamiri, S. S. et al. Automated inference of Boolean models from molecular interaction maps using CaSQ. Bioinformatics 36, 4473–4482 (2020).
Data analysis revealed 1625 DEG from three different datasets of expression data in Sjögren patients. Pathway enrichment analysis using GSEA and Reactome database identified 21 common enriched pathways. These pathways along with literature-based search were used as the starting point for the building of the Sjögren molecular interaction map (MIM). The first version of the map consists of 187 biomolecules organized in 16 pathways (5 data-driven and 11 literature -mined) and has served as a template for the inference of a discrete computational model. The model comprises 111 biomolecules and 130 interactions.
Acknowledgements: I express my gratitude to the NECESSITY consortium members for the materials they made available to me, which enabled me to carry out this research. I would also like to thank the doctoral school SDSV of Paris-Saclay University and Genopole for financing respectively my PhD studies and formations I followed.
Disclosure of Interests: Sacha E Silva-Saffar: None declared, Jacques-Eric Gottenberg BMS, Pfizer, Lilly, Abbvie, AstraZeneca, Gilead, Galapagos, MSD, Roche-Chugai, Sanofi, UCB, Michele Bombardieri: None declared, Divi Cornec: None declared, Jacques-Olivier Pers: None declared, Marta Alarcon-Riquelme: None declared, Philippe MOINGEON Sanofi, Stallergenes, Servier, Michael Barnes: None declared, Sandra Ng: None declared, Wan-Fai Ng GlaxoSmithKline, MedImmune, Novartis and BMS, Abbvie, Resolves Therapeutics, Nascient, Xavier Mariette Sanofi, Servier, BMS, Gaetane Nocturne Amgen, Novartis, Anna Niarakis Sanofi.