

Background: Rheumatoid arthritis (RA) is a complex heterogenous autoimmune disease where 40% of patients are non-responsive to individual agents. Therefore, there is an urgent need to better strategise treatment through understanding the disease molecular mechanisms and its response to therapy. The biopsy driven clinical trial, R4RA, demonstrated that patients with low B cell signatures responded more to tocilizumab compared to rituximab[1]. Previous molecular analysis of the R4RA synovial biopsies using RNA-seq demonstrated immune response signatures which predicted future response to rituximab or tocilizumab as well as a stromal signature in refractory patients to both treatments[2]. However, how this translates to functional protein levels and active signalling pathways is not known and there is a paucity of phosphoproteome studies in RA.
Objectives: We aimed to deepen our understanding on the potential pathways driving the different RA pathotypes and response to rituximab or tocilizumab in late-stage RA patients from the R4RA trial using phosphoproteome and total proteome analysis.
Methods: 33 baseline synovial biopsies before treatment from the trial were analysed using label-free mass spectrometry. All three pathotypes were represented: lymphoid (n=21), myeloid (n=7) and fibroid (n=5). 15 patients of the synovial biopsies went on to receive rituximab where 4 were responders and 11 were non-responders as determined by Disease Activity Score (DAS)-28-ESR < 3.2 and an improvement of > 1.2 after 16 weeks of treatment. The remaining 18 were treated with tocilizumab where 10 were responders and 8 were non-responders as determined by the same criteria as rituximab.
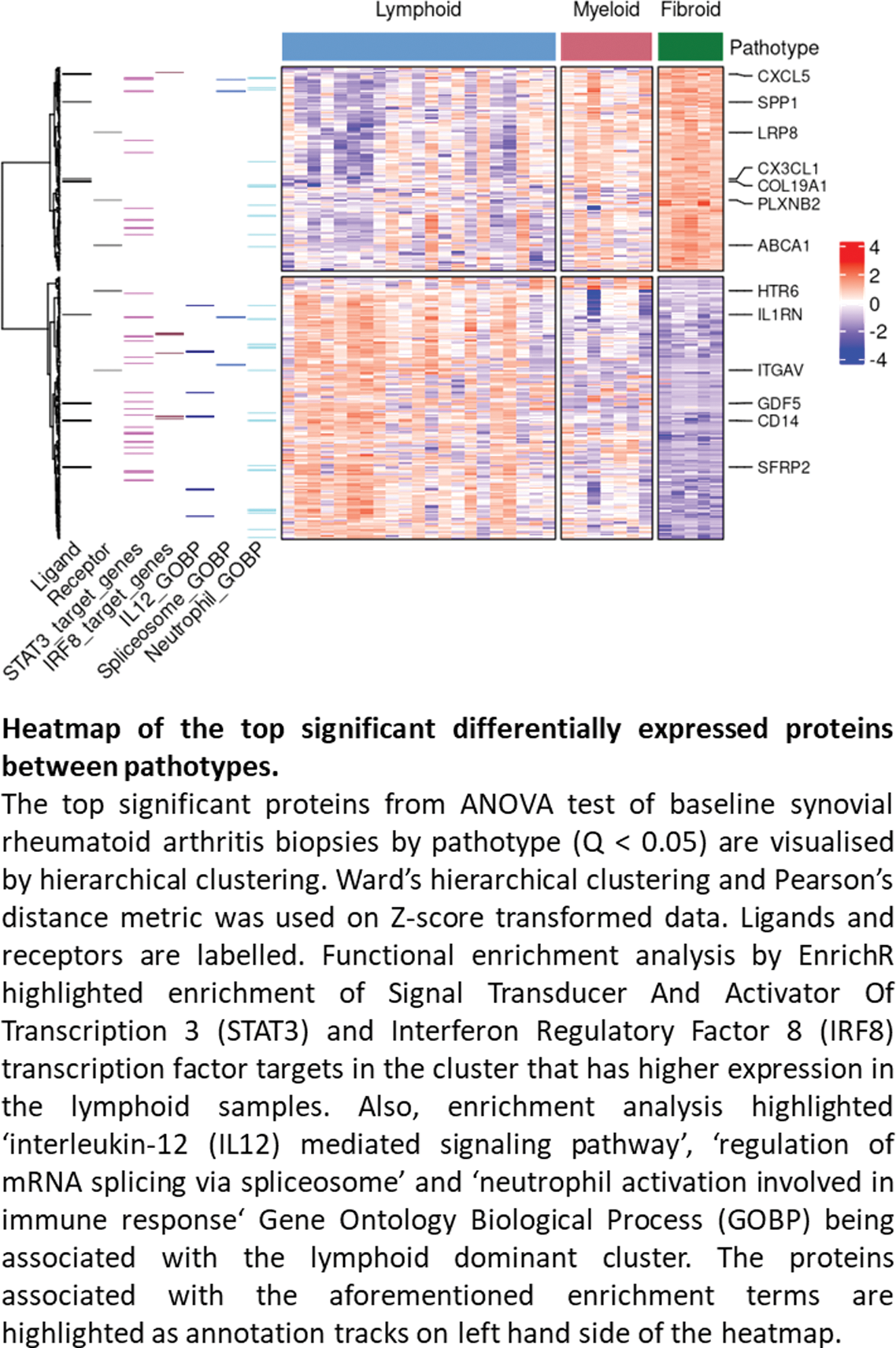
Results: 5,489 proteins and 12,989 phosphosites were quantified. The different pathotypes of the synovial biopsies were a key determinant of both phosphoproteome and proteome profiles. Immunological activity corresponded to the pathotypes with lymphoid being the most active, followed by myeloid and fibroid. For instance, the lymphoid pathotype had upregulated B cell related kinase activity, such as Mammalian Target of Rapamycin (MTOR, p=0.001), and enrichment of proteins regulated by interferon-regulated factor 8 (IRF8, q=0.02) and Signal Transducer and Activator Of Transcription 3 (STAT3, q=0.0003) (Figure 1). Characterisation of rituximab responders and non-responders using baseline synovial biopsies highlighted the enrichment of B cell related kinases in rituximab responders before treatment (BLK, LYN, SYK p=0.001 and MTOR p=0.007). For tocilizumab, there was a predicted enrichment of Mitogen Activated Protein Kinase (MAPK) activity in tocilizumab responders before treatment (e.g MAPK2K6 p=0.034 and MAP2K3 p=0.035), which may reflect the reciprocal interactions of MAPK and interleukin 6 (IL6) receptor signalling.
Conclusion: This phosphoproteome study demonstrates the distinct baseline phosphoproteome profiles for each RA pathotype and the pre-treatment signalling pathways associated with drug response. Therefore, it highlights the importance of exploring the signalling network to improve personalised therapeutic strategies for managing RA.
REFERENCES: [1] Humby et al Lancet 2021 397 (10271): 305-217.
[2] Rivellese et al Nat Med 2022 28 (6): 1256-1268.
Acknowledgements: The R4RA study was supported by the UK National Institute of Health Research (grant reference: 11/100/76) and core work associated with this project was supported by Versus Arthritis (Experimental Arthritis Treatment Centre, grant number 20022) and Barts and The London School of Medicine and Dentistry charity (grant number 523/819), MRC and Arthritis Research UK (ARUK) by their joint funding of Maximizing Therapeutic Utility in Rheumatoid Arthritis (MATURA) (grant numbers MR/K015346/1 and 20670 respectively), NIHR (grant 131575) and MRC TRACT-RA (MR/ V012509/1). This work acknowledges the support of the National Institute for Health Research Barts Biomedical Research Centre (NIHR 203330). The work of PC and VR is funded by Cancer Research UK (C15966/A24375 and C16420/A18066).
Disclosure of Interests: None declared.