

Background: VEXAS syndrome is an acquired autoinflammatory disorder secondary to somatic mutations of UBA1 in bone marrow progenitors. Its phenotypic expression combines inflammatory features, cytopenia, and myelodysplastic traits, and the diagnosis relies on UBA1 sequencing. Situations giving rise to suspicion of VEXAS syndrome have broadened, but it remains a relatively rare disease. Consequently, it seems important to select patients in whom genetic analysis is most relevant. Since the presence of bone marrow vacuoles is greatly significant in the diagnosis of VEXAS syndrome [1], we hypothesized that the vacuolization of circulating immature myeloid cells, and/or the morphology of blood mature myeloid cells could help for distinguishing VEXAS syndrome from differential diagnoses. In contrast to bone marrow aspiration, blood smear analysis is a minimally invasive examination which could be used to screen large populations for VEXAS syndrome.
Objectives: We investigated the association between VEXAS syndrome and i) the vacuolization of circulating immature myeloid cells, and ii) the morphology of blood mature myeloid cells, and we thereafter assessed their diagnostic performances for VEXAS syndrome.
Methods: To test for the vacuolization of circulating immature myeloid cells, we compared the proportion of vacuolated cells, the number of vacuoles and the vacuoles’ area between VEXAS and UBA1-WT (‘VEXAS-like’ UBA1-wild type) patients who had circulating immature myeloid cells on blood smears within 3 months of the UBA1 sequencing [2]. To test for the morphology of blood mature myeloid cells, we used deep learning analysis of images of leukocytes in a two-step procedure: first, we used self-supervised contrastive learning to train convolutional neural networks to translate leukocytes images into lower dimensional encodings, then we employed support vector machine to predict patients’ condition based on those leukocytes’ encodings [3]. We compared the images of leukocytes from 3 conditions: VEXAS syndrome, MDS (myelodysplastic syndrome), and UBA1-WT ‘VEXAS-like’ patients.
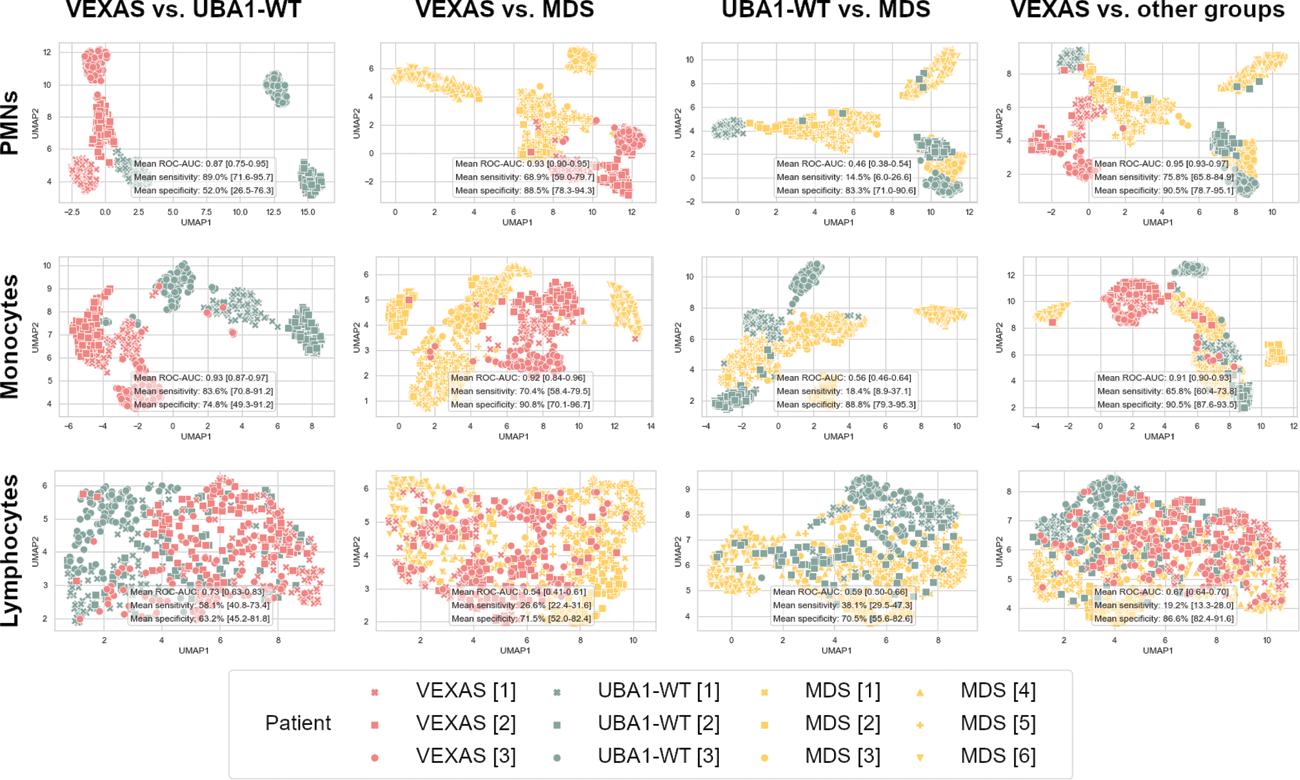
Results: We analysed 99 blood smears including immature myeloid cells (15 among VEXAS patients, 84 among UBA1-WT patients). Neither the proportion of vacuolated circulating immature cells (p=0.36) nor the number of vacuoles per vacuolated cell (p=0.49), nor the surface area of vacuoles per vacuolated cell (p=0.34) differed between the VEXAS and UBA1-WT groups. The deep learning analysis of 33 757 images of neutrophils and monocytes enabled us to distinguish VEXAS patients from both UBA1-WT and MDS patients with ROC-AUC ranging from 0.87 to 0.95 (Figure 1). In contrast, the analysis of lymphocytes images (used as a negative control) was not associated with the diagnosis of VEXAS syndrome.
Conclusion: The invasive nature of bone marrow aspiration prevents its use for a large screening purpose. In contrast, the blood smear analysis is more suitable for widespread screening. The similar vacuolization of circulating immature myeloid cells between VEXAS and ‘VEXAS-like’ UBA1-WT patients prevent its use for a screening purpose and is explained by the main bloodstream passage of late precursors, in which the vacuolization is already similar in bone marrow in both cases. At the opposite, the deep learning analysis of blood mature myeloid cells is a promising tool for large VEXAS screening, but it will probably require a calibration in each centre.
REFERENCES: [1] Lacombe V, Prevost M, Bouvier A, et al. Vacuoles in neutrophil precursors in VEXAS syndrome: diagnostic performances and threshold. Br J Haematol 2021;195:286–9.
[2] Lacombe V, Genevieve F, Chabrun F. Vacuoles in circulating immature myeloid cells in VEXAS syndrome and comparison with UBA1 -wild type patients. American J Hematol 2023;ajh.27192.
[3] Chabrun F, Lacombe V, Dieu X, et al. Accurate stratification between VEXAS syndrome and differential diagnoses by deep learning analysis of peripheral blood smears. Clin Chem Lab Med. 2023;61:1275–9.
Median encodings of leukocyte images of patients in VEXAS (red), UBA1-WT (green) and MDS (yellow) groups, after UMAP dimension reduction.
Acknowledgements: NIL.
Disclosure of Interests: None declared.