

Background: The loss of tolerance to nuclear antigens characterizes systemic lupus erythematosus (SLE), where CD4 T cells play a pivotal role in aiding B cells that produce autoantibodies. Despite this, the extent of aberrant T cell plasticity in SLE remains incompletely understood.
Objectives: Our aim is to investigate T cell plasticity in SLE patients and characterize its association with SLE disease phenotype, utilizing large-scale peripheral blood T cell RNA-seq data from the Immune Cell Gene Expression Atlas from the University of Tokyo (ImmuNexUT) [1,2].
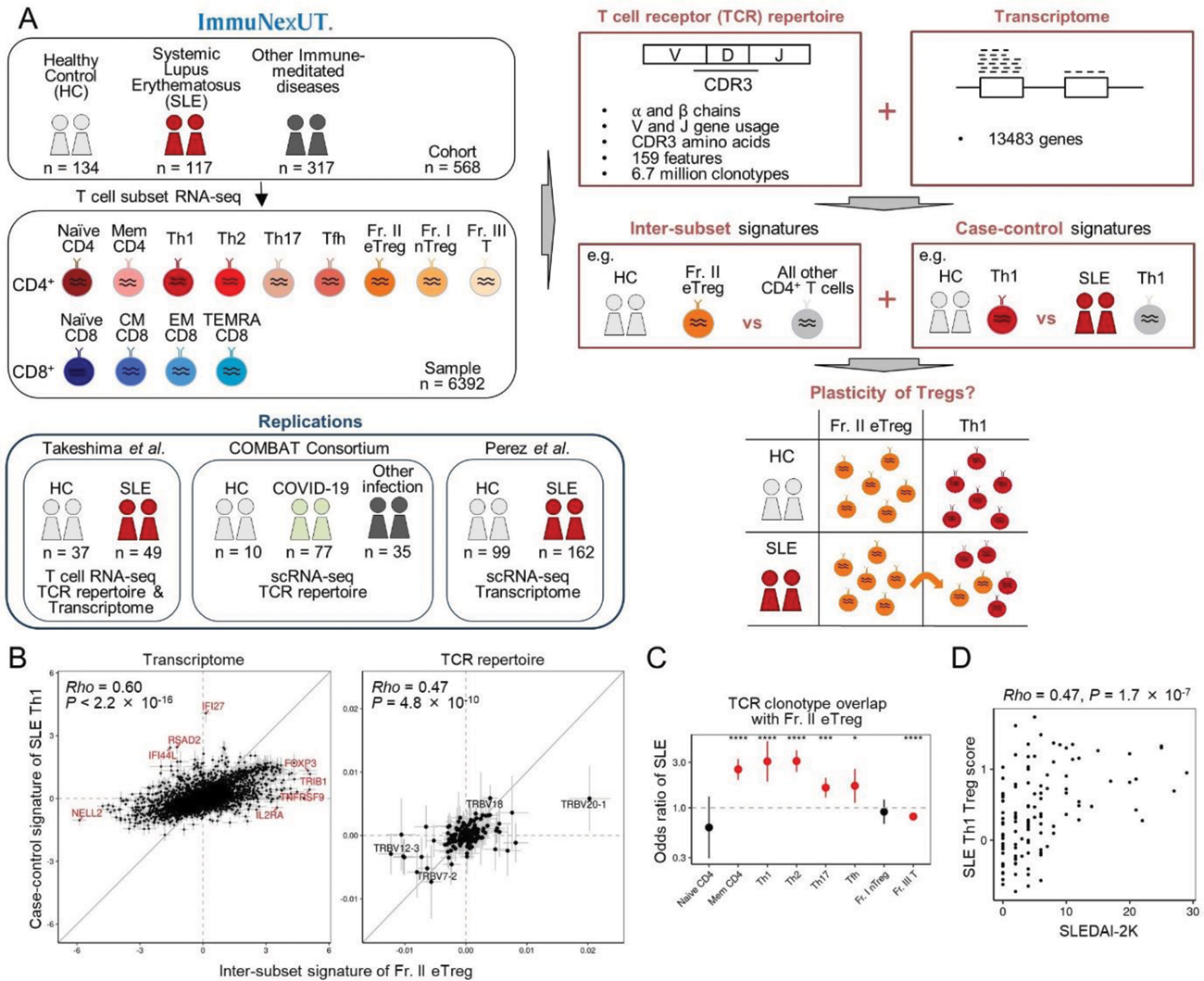
Methods: We recruited 134 healthy controls (HC), 117 SLE patients, and 317 patients with 9 other immune-mediated diseases (Figure 1A). Nine CD4 T cell subsets and four CD8 T cell subsets were fine-sorted from each patient for RNA-seq analysis, resulting in 6392 T cell samples. Utilizing T cell receptor (TCR) repertoire analysis, we extracted 159 TCR repertoire features, encompassing V and J gene usage and CDR3 amino acid patterns from both α and β chains. We characterized inter-subset T cell signatures by comparing the features of TCR repertoire sequences or transcriptomes with those of other T cell subsets, such as Fr. II eTreg in comparison to other CD4 T cell subsets. Case-control signatures of T cell subsets were analyzed, comparing SLE (and other diseases) to HC. Subsequent correlation analysis provided insights into aberrant T cell differentiation in patients in vivo . We assessed the TCR clonotype overlap between CD4 T cell subsets using logistic mixed effect analysis. With transcriptomic Fr. II eTreg inter-subset signatures, we developed the “Th1 Treg score” to quantify the Treg-likeliness of each Th1 sample. The clinical relevancy of Th1 Treg score was evaluated. The identified signatures and correlations were validated for reproducibility using publicly available (single cell) RNA-seq datasets.
Results: Analysis of inter-subset signatures within T cell subsets unveiled distinctive gene signatures (for example, FOXP3 , TRIB1 , and TNFRSF9 for Fr. II eTreg, and TBX21 and CXCR3 for Th1). Additionally, characteristic TCR CDR3 amino acid usages were identified, such as highly hydrophobic amino acids in Fr. II eTreg and acidic amino acids in Th1. Correlation analysis showed the most robust association between the inter-subset Fr. II eTreg signature and the case-control signature of SLE Th1 (Figure 1B). Further validation through TCR repertoire clonotype overlap analysis affirmed the heightened plasticity in SLE compared to healthy controls, particularly between Fr. II eTreg and effector CD4 T cells, including Th1 (Figure 1C, odds ratio = 3.1, P = 9.1 × 10 -6 in Th1). Evaluation of Th1 Treg scores in SLE patients revealed a positive correlation with SLEDAI-2K score (Figure 1D). Moreover, Th1 Treg scores were elevated in active SLE patients presenting with fever or nephritis.
Conclusion: Our study unveils plasticity between regulatory T cells and effector Th1 cells in SLE, shedding new light on SLE pathophysiology. This finding holds potential as a biomarker reflecting disease activity.
REFERENCES: [1] Ota, M. et al. Dynamic landscape of immune cell-specific gene regulation in immune-mediated diseases. Cell 184,3006-3021.e17 (2021).
[2] Nakano, M. et al. Distinct transcriptome architectures underlying lupus establishment and exacerbation. Cell 185, 3375-3389.e3321 (2022).
Plasticity of SLE regulatory T cells. (A) Study overview. 9 other immune-mediated diseases are systemic sclerosis, idiopathic inflammatory myopathy, rheumatoid arthritis, ANCA-associated vasculitis, Behçet’s disease, mixed connective tissue disease, adult-onset Still’s disease, Sjögren’s syndrome, and Takayasu arteritis. (B) The correlation of Inter-subset signature of Fr. II eTreg and Case-control signature of SLE Th1. (C) TCR clonotype overlap analysis with Fr. II eTreg in SLE. (D) The correlation between SLEDAI-2K score and SLE Th1 Treg score.
Acknowledgements: We thank all the study participants and all the members of the recruitment sites for the collection of data.
Disclosure of Interests: Keishi Fujio receives consulting honoraria from Chugai Pharmaceutical., Keishi Fujio receives research support from Chugai Pharmaceutical., Yasuo Nagafuchi belonged to the Social Cooperation Program, Department of Functional Genomics and Immunological Diseases, supported by Chugai Pharmaceutical., Masahiro Nakano: None declared, Mineto Ota belonged to the Social Cooperation Program, Department of Functional Genomics and Immunological Diseases, supported by Chugai Pharmaceutical., Hiroaki Hatano: None declared, Takahiro Itamiya belong to the Social Cooperation Program, Department of Functional Genomics and Immunological Diseases, supported by Chugai Pharmaceutical., Tomohisa Okamura belong to the Social Cooperation Program, Department of Functional Genomics and Immunological Diseases, supported by Chugai Pharmaceutical., Kazuyoshi Ishigaki: None declared.