

Background: Pain as a symptom of knee osteoarthritis (OA) is a formidable problem for which safe, effective pharmacological treatments are urgently needed. A critical gap hampering progress in our understanding of the mechanisms underlying joint pain is the lack of a complete characterization of the sensory innervation of the healthy and OA-affected knee. We also lack a precise description of molecular and cellular changes in dorsal root ganglia (DRG) neurons that innervate the knee.
Objectives: We propose a holistic approach for precisely documenting the sensory innervation of the human and murine knee and the dynamic neuroplastic changes occurring with OA, as well as to elucidate the molecular changes that drive this neuroplasticity.
Methods: We use state-of-the-art imaging techniques combined with transcriptomics for (1) constructing 3D models of knee sensory innervation; (2) composing a cell atlas in which knee afferents are transcriptionally profiled at a single cell resolution, and (3) documenting the nerve-joint cell “interactome” at the transcriptional level. Mouse studies : Using clearing-enabled lightsheet microscopy (Zeiss LightSheet 7), we construct high-resolution 3D anatomical models of joint innervation in Na V 1.8 tdTomato reporter mice. We construct molecular maps of knee-innervating neurons in the DRG using spatial transcriptomics. This will identify precisely which DRG neurons innervate the healthy and diseased knee and will describe their molecular phenotypes. Human studies : We are characterizing the sensory innervation of human knees and matched DRGs, using post mortem knee-DRG samples from (1) healthy knees, age 20-40 (n=15/sex); (2) knees from donors > 70 (n=15/sex), in which > 80% exhibits OA pathology. Knee tissues are collected in a standardized fashion, including synovium, osteochondral plugs (OCP), meniscus, fat pad, cruciate ligaments. In each tissue, we perform (1) histopathology; (2) IHC for sensory nerves and blood vessels; (3) bulk RNAseq; (4) single nuclei multiome; (5) spatial transcriptomics. Matched DRGs will be used for bulk RNAseq to identify differentially expressed genes between groups and enable ligand-receptor analysis. Data from joint tissues will be mined for mediators of neuronal plasticity and pain, and their spatial expression correlated with changes in innervation patterns. Nerve-synovium interactome: We are comparing patient report of OA knee pain at the time of total knee replacement to synovial histology, innervation, and scRNAseq. We also reconstruct the cellular interactome between synoviocytes and DRG neurons in mouse models using scRNAseq of matched synovium and DRGs.
Results: Mouse studies : We successfully achieved immunolabeling and light sheet imaging of tdTomato protein in Na V 1.8;tdTomato mice, showing robust imaging of nerves in intact mouse knees and L5 DRG (Figure 1), setting the stage for detailed anatomical studies. Human studies : An initial analysis using OCP from mild OA (grade 1.5, Modified Outerbridge) and advanced OA donors (grade 3) at 4 tibial sites revealed that PGP9.5+ and CGRP+ neuronal signals were present in osteochondral channels that approach (Figure 2a) or breach the tidemark (Figure 2b-d). Endothelial markers CD31and CD34 were also observed. Concomitantly, single nuclei multiome analysis revealed 12 clusters in osteochondral tissue from an OA (72M, grade 4) donor. One of these subsets (cluster 5) expressed genes involved in synaptic plasticity.
Conclusion: Our integrative approach to mapping the nerve-joint interactome of mouse and human knees aims to document their sensory innervation with concomitant molecular changes in the DRG and joint tissues. Preliminary results suggest feasibility of the approach as a prelude to systematic analysis. We expect that molecular phenotyping of DRG neurons and the nerve-joint interactome will identify mediators and their receptors that contribute to joint pain, neuroplasticity, and joint integrity. This will result in comprehensive databases of the neuro-articular environment, which can be mined to identify pharmacologically tractable targets for inhibiting pathological joint neuroplasticity and pain.
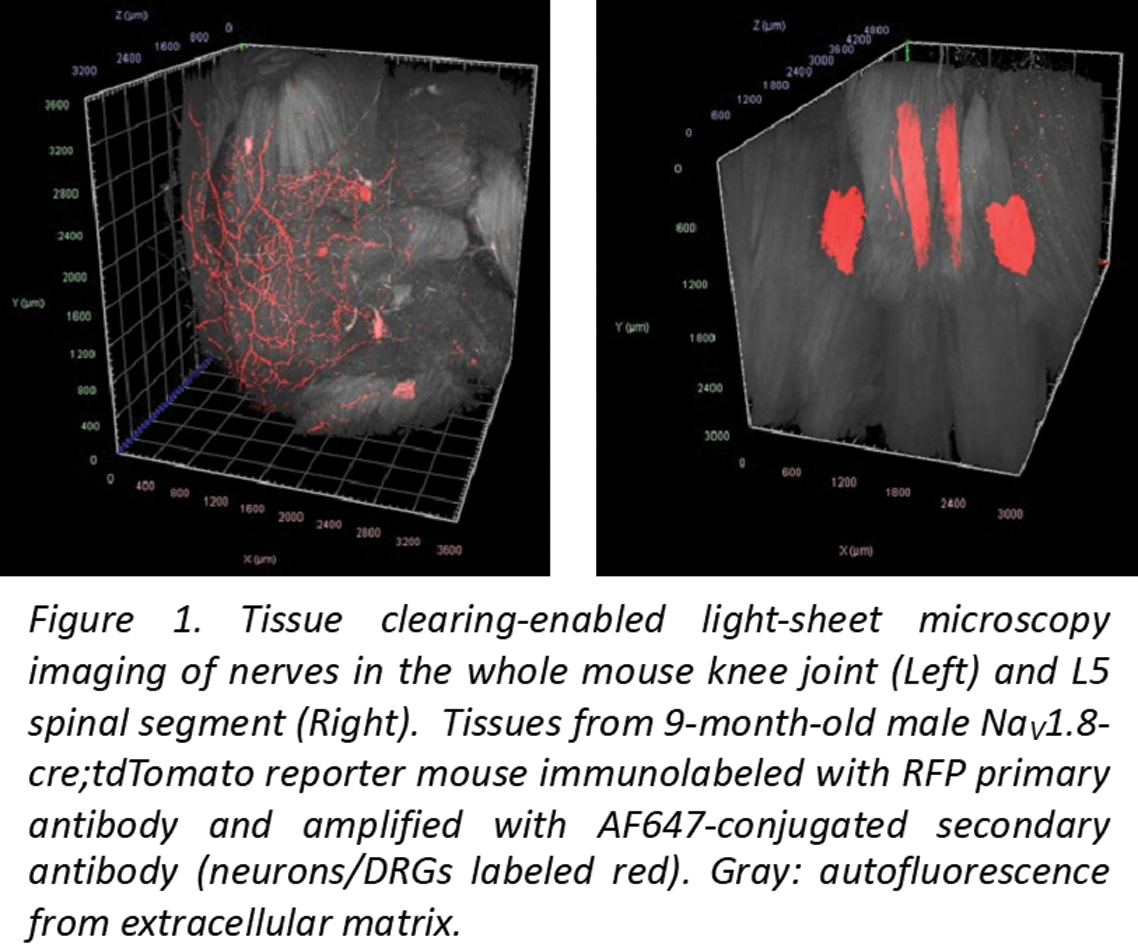
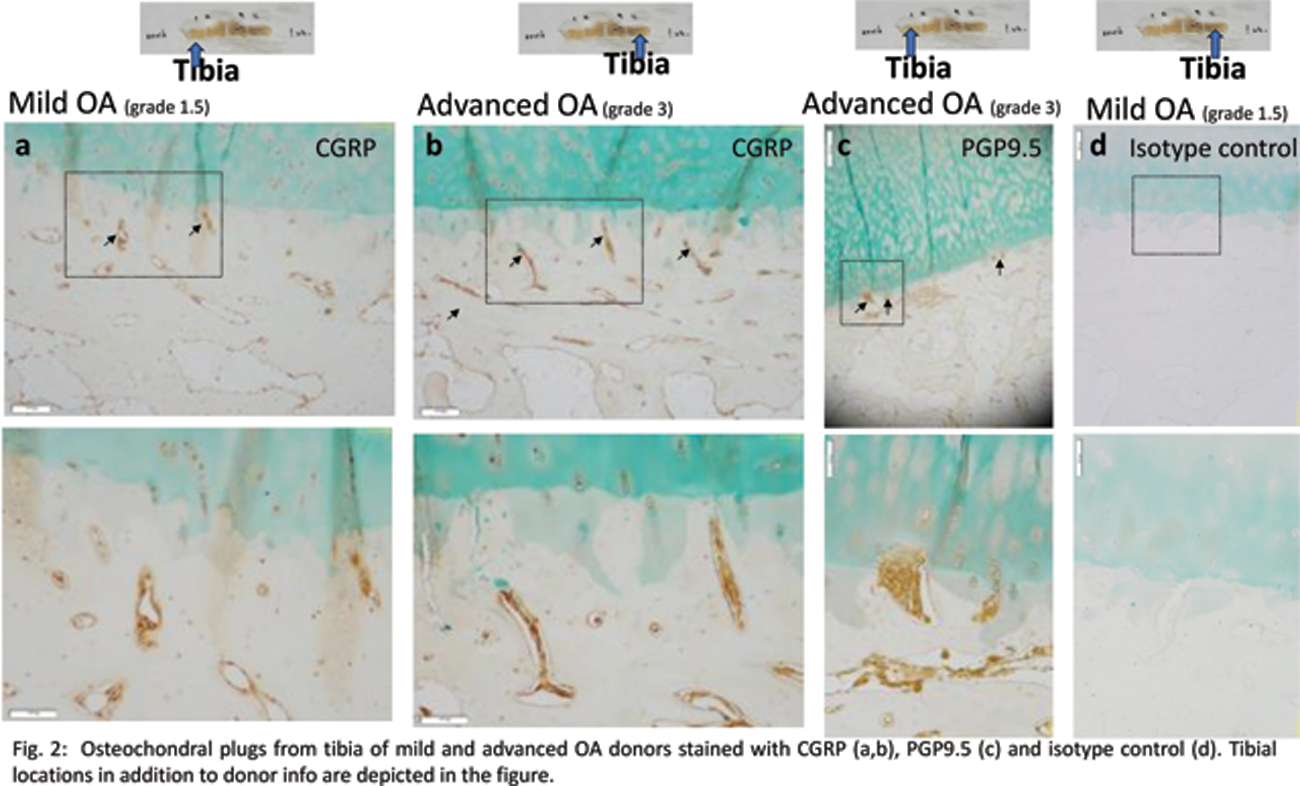
REFERENCES: NIL.
Acknowledgements: The study is funded by UC2AR082186, part of the NIAMS Rejoin Consortium.
Disclosure of Interests: Anne-Marie Malfait Orion, Merissa Olmer: None declared, Alia Obeidat: None declared, Jun Li: None declared, Hannah Swahn: None declared, Michael Newton: None declared, Frank Ko: None declared, Bella Mehta: None declared, Miguel Otero: None declared, Tristan Maerz: None declared, Dana Orange: None declared, Rachel Miller: None declared, Richard Miller: None declared, Martin Lotz: None declared.