

Background: Systemic sclerosis (SSc) is a heterogenous disease characterised by autoimmunity, vasculopathy, and fibrosis.
Objectives: This study aimed to characterize the peripheral immunome associated with active disease in systemic sclerosis. Our findings will shed light into SSc disease pathogenesis. Furthermore, our findings will provide an opportunity to develop potential theragnostic tool to evaluate active disease in early stage SSc.
Methods: Peripheral blood mononuclear cells (PBMC) were collected from SSc patients with early disease (<5 years from non-Raynaud’s phenomenon; n=13 active, n=12 inactive), and age-and-sex matched healthy controls (n=10). Active disease was defined by composite EUSTAR-activity index score ≥2.5. Single-cell proteomic data was acquired using mass cytometry (CyTOF), with a panel of 77 unique markers. The CyTOF data was analysed using the Extended Polydimensional Immunome Characterization (EPIC) platform, which is an unsupervised machine learning algorithm to characterize immune cell subset perturbations. Using cell frequencies of the identified cell subsets, we built multiple logistic regression models to classify active and inactive diseases. Lastly, a correlation network between nodes was constructed for active SSc and inactive SSc by performing pairwise correlations between each node pair (absolute correlation coefficient cut off > 0.6).
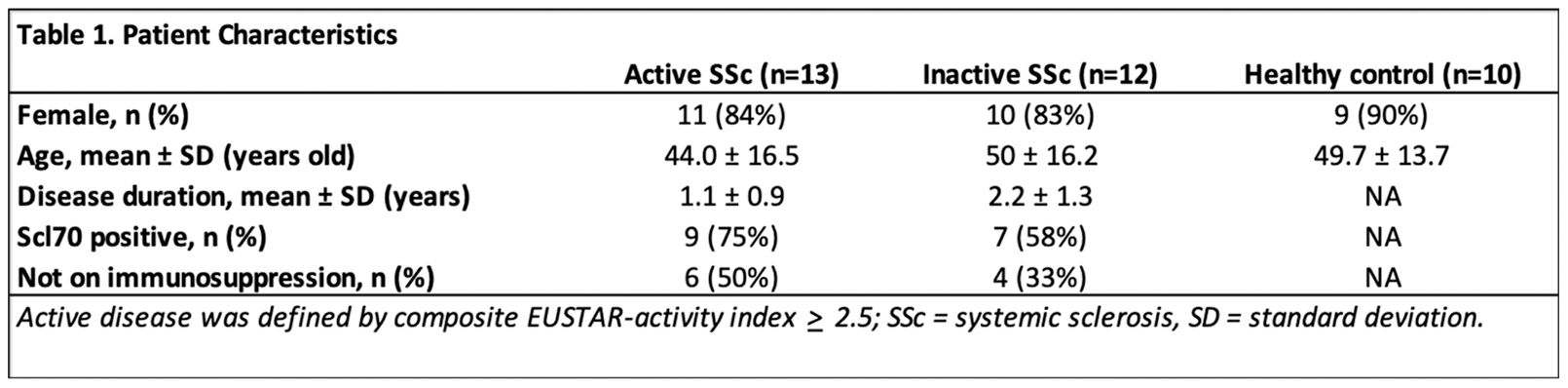
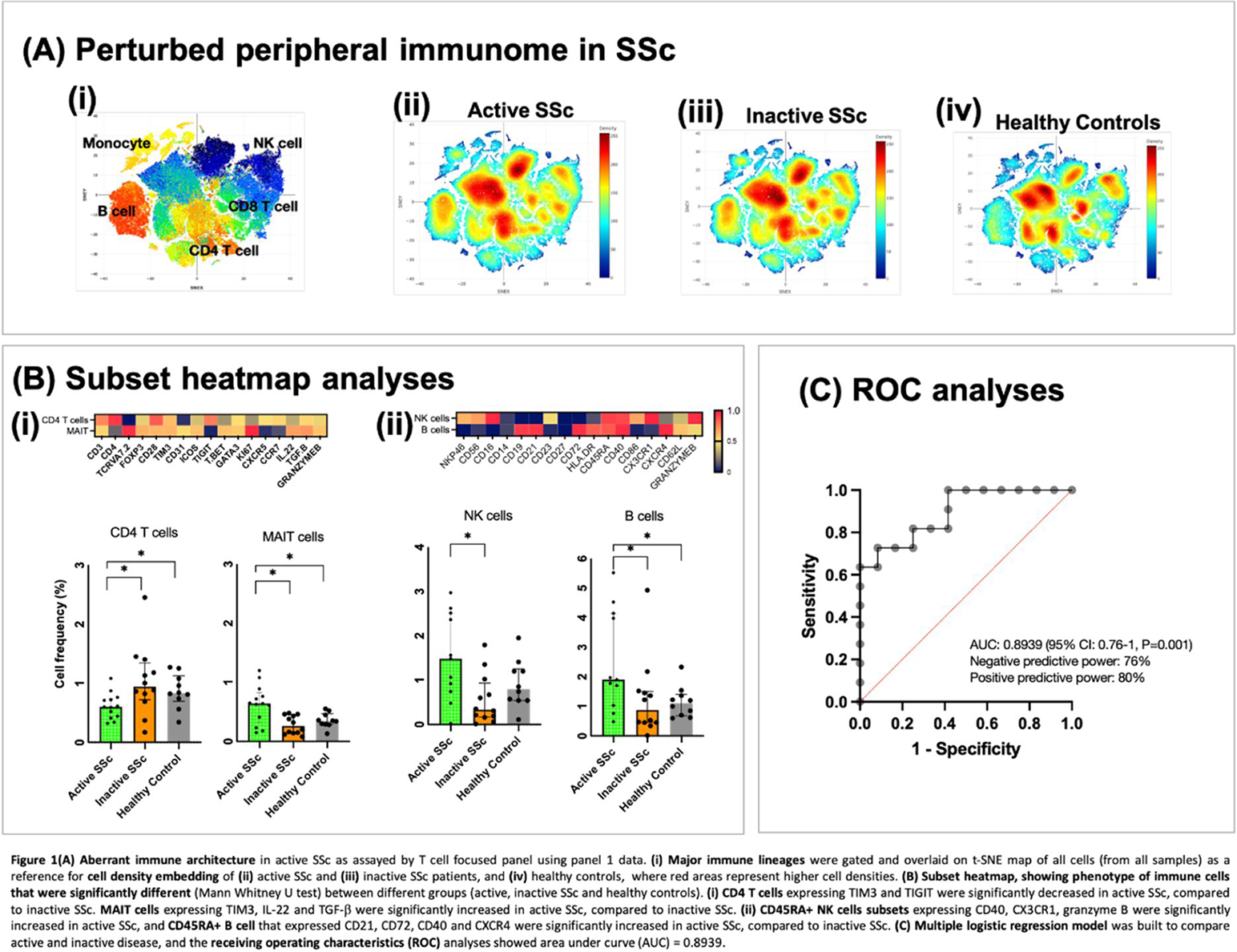
Results: Patient characteristics are summarized in Table 1. Dysregulations in various immune cell lineages were observed in the peripheral immunome of active SSc, as compared to inactive SSc and healthy controls (Figure 1A). CD4 T cells expressing TIM3 and TIGIT (known immune checkpoint) were significantly decreased in active SSc, compared to inactive SSc (Figure 1Bi). MAIT cells expressing TIM3, IL-22 and TGF-β were significantly increased in active SSc, compared to inactive SSc (Figure 1Bi). CD45RA+ NK cell subsets expressing CD40, CX3CR1, granzyme B were significantly increased in active SSc; whereas CD45RA+ B cells that expressed CD21, CD72, CD40 and CXCR4 were significantly increased in active SSc, compared to inactive SSc (Figure 1Bii). By using cell frequencies of the identified immune cell subsets, we built a multiple logistic regression model and the Receiver Operating Characteristic (ROC) curve analysis showed an area under the curve (AUC) of AUC: 0.8939 (95% CI: 0.76-1, P=0.001, with negative predictive power: 76% and positive predictive power: 80% (Figure 1C). Immune cell network from SSc patients with inactive disease showed more negatively connected edges compared to network from SSc patients with active disease. In addition, the modularity of the immune cell network is also higher in the inactive SSc than in the active SSc. A subset of cells had more connections (higher degree of centrality) in the inactive SSc, such as the CD4 T cells that expressed immune checkpoints. However, other subset of cells that formed module on the edge in the inactive disease state had become more distributed in the network with increased degree of centrality in the active SSc, such as CD45RA+ NK cells that expressed CD40, CX3CR1 and granzyme B, as well as CD45RA+ B cells expressing CD21, CD72, CD40 and CXCR4.
Conclusion: We identified dysregulated immune cell subsets with distinct expression of activation markers, immune checkpoints, and chemokine receptors, suggesting perturbed immune cell trafficking and regulatory mechanisms. Correlation network analysis suggests that CD4 T cells with known immune checkpoints may have limited ability to interact with other cell types in active disease, hence affecting peripheral immune tolerance. The identification of dysregulated immunonome in active SSc will allow for more focused scientific questions, such as investigating the functional significance of these immune cell subsets and their roles in the disease pathogenesis. With further validation studies, the identified immune signatures could potentially be used as theragnostic tools to evaluate disease activity in SSc.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.