

Background: Knee osteoarthritis (KOA) is the most common form of arthritis. The infrapatellar fat pad (IFP) in the knee is responsible for a variety of functions including shock absorption and structural support. The exact contributions of the IFP to knee OA is not well understood. Currently, the exact cell populations within the IFP and its role in KOA pathology remain to be characterized.
Objectives: This study aims to identify distinct cell types, subsets, and transcriptomic profiles within the IFP, and define changes due to KOA and obesity status using single-nucleus RNA sequencing (snRNA-seq), spatial sequencing and bioinformatic analysis. To the best of our knowledge, this is the first cellular and transcriptomic atlas of knee OA IFP.
Methods: IFP was obtained from late-stage KOA patients [KL grades III/IV; n=15] during total knee replacement. Healthy control donor IFPs (n=6; post-mortem, obtained < 4h after death) were obtained from individuals without known musculoskeletal disease. Nuclei underwent snRNA-seq on an Illumina NextSeq 550 using the 150bp high output sequencing kit. Data was processed using Cell Ranger and clusters were annotated using canonical markers while differential gene expression testing determined a gene signature. IFP [n=10] were spatially sequenced using Visium CytAssist techniques and analyzed using Seurat.
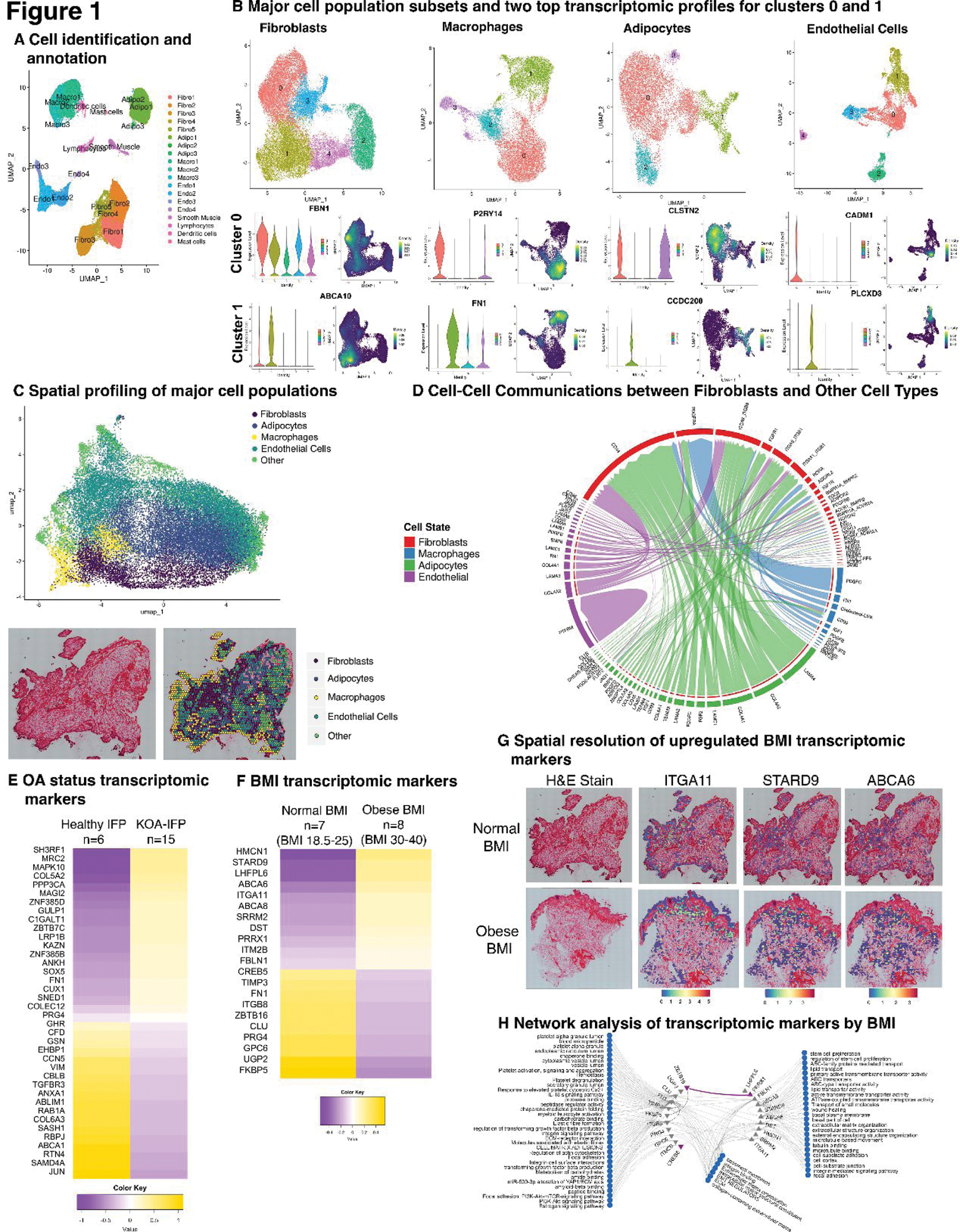
Results: Using snRNA-seq, 73,808 nuclei were analyzed from 21 IFP samples. Cluster analysis revealed eight cell types present, with major populations being fibroblasts, macrophages, adipocytes, and endothelial cells (Figure 1A). Independent cluster analysis of each major cell type elucidated multiple subsets, each with a unique transcriptomic profile (Figure 1B). Spatial sequencing confirmed fibroblasts, macrophages, adipocytes, and endothelial cells are the major cell types present and are distributed across the IFP (Figure 1C). We also spatially resolved transcriptomic markers of each fibroblast subset across the IFP. Putative ligand-receptor interaction analysis uncovered multiple cell communications received by fibroblasts (Figure 1D). Regardless of directionality, each major cell type also has a unique outgoing signalling pattern, indicating the potential for various inter-cell communications. Fibroblasts were the predominant cell type based on proportion of nuclei; thus, downstream analysis focuses on fibroblasts. When comparing fibroblasts between KOA-IFPs (n=15) and healthy IFPs (n=6), there were no variations in the presence of cell subsets, however moderate differences in subset proportions were observed. Within KOA compared to healthy IFPs, 38 transcriptomic markers were differentially expressed: 20 upregulated and 18 downregulated genes (Figure 1E). Analysis of obese (n=8) vs normal BMI (n=7) KOA-IFP revealed no difference in presence or proportion of fibroblast subsets. However, 21 transcriptomic markers were differentially expressed: 10 upregulated and 11 downregulated genes (Figure 1F). Of the upregulated genes, 3 were significantly upregulated within spatial analysis (Figure 1G). GO and Pathdip analysis identified enriched biological processes and pathways linked to identified transcriptomic markers, suggesting differences in fibroblast function within KOA-IFP based on BMI. (Figure 1H).
Conclusion: To the best of our knowledge, using snRNA-seq, spatial sequencing and bioinformatics analysis we have created the first cellular and transcriptomic atlas of knee OA IFP. We have revealed transcriptomic differences within fibroblasts based on KOA and obesity status. Ongoing experiments will further investigate functions of IFP fibroblasts to determine the implications of differences in the transcriptomic profiles between obese versus normal weight KOA patients.
Cellular and transcriptomic profiles of knee OA IFP using snRNA-seq and spatial sequencing.
REFERENCES: NIL.
Acknowledgements: This work is funded by The Arthritis Society of Canada Strategic Operating Grant, Tony and Shari Fell Platinum Chair in Arthritis Research and Canada Research Chairs Program.
Disclosure of Interests: None declared.