

Background: Osteoarthritis (OA) is a prevalent condition among people of advanced ages and presents with pain, stiffness, and in severe cases disability. Currently, there are no approved disease-modifying drugs for OA and pharmacotherapeutic options are limited to symptomatic relief with analgesics and anti-inflammatories. In addition to variable and incomplete response to treatment, patients present a variable natural history and progression suggesting heterogeneity within OA.
Objectives: To characterise peripheral immunome dysregulations as a result of local joint inflammation in knee OA, and to identify unique peripheral immune signatures for patient stratification into different OA subtypes.
Methods: We employed mass cytometry time-of-flight (CyTOF) to investigate peripheral blood mononuclear cells (PBMCs) obtained from patients with knee OA and healthy controls (HCs). 57 patients with knee OA and 33 age, sex, and ethnicity-matched HCs were enrolled in this study. Two CyTOF marker panels that cover major innate and adaptive immune cell lineages were designed to interrogate these samples to capture broad immune lineage changes. The resulting single-cell proteomic CyTOF data was analysed using in-house bioinformatic pipeline to identify peripheral immunome dysregulations. Further statistical and machine learning algorithms were applied to the immune clusters generated to extract unique peripheral immune signatures for patient stratification into different disease subtypes.
Results: Broad dysregulations in both the innate and adaptive compartments of the peripheral OA immunome were observed. We observed an increase in monocyte, NK, and various T cell subsets in OA as compared to HC peripheral immunome. Using a combination of statistical analyses and machine learning models, we identified 10 immune clusters in which we applied unsupervised K-means clustering which revealed three OA subtypes with distinct clinical phenotypes and corresponding peripheral immune signatures. We identified an inflammatory subtype (Group 3) where patients present clinically with greater pain and stiffness, and poorer function. Patients with this inflammatory subtype have a peripheral immune signature enriched with inflammatory immune subsets such as various monocyte and effector T/T-like immune subsets. Another OA subtype (Group 2) with an immune-modulated profile was also identified where patients report less pain and stiffness. The peripheral immune signature of this group of patients suggests increased immune regulation as seen in the relative increase in Treg and exhausted Th subsets in the peripheral immunome.
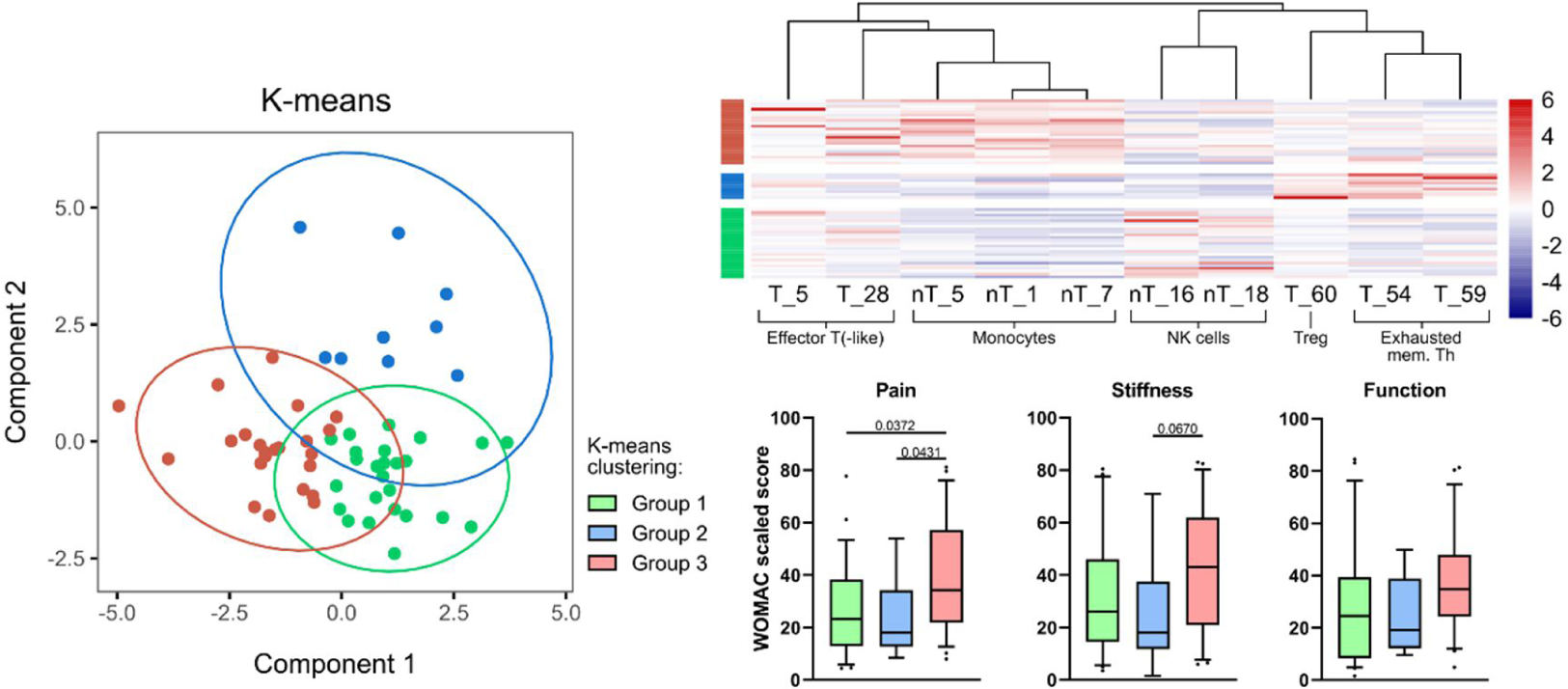
Conclusion: In conclusion, our application of statistical and machine learning approaches in the analysis of high-dimensional immunomic CyTOF data yielded a set of 10 immune clusters that can stratify patients into three clinical subtypes with distinct immunological differences. Findings from our study have clinical applications as a promising precision medicine tool in the comprehensive management of OA where pharmacotherapeutic decisions can be made based on a patient’s peripheral immunophenotype. For example, our identification of an inflammatory subtype may benefit from early immunomodulatory efforts to arrest the inflammatory process and prevent disease progression. All in all, our high-dimensional approach in the analysis of immunological data of patients with OA uncovered clinically significant subtypes and insights into disease pathogenesis.
REFERENCES: NIL.
Acknowledgements: Alfred.
Disclosure of Interests: Alfred Chia: None declared, Ahmad Lajam: None declared, Gladys Ang: None declared, Martin Wasser: None declared, Ying-Ying LEUNG AbbVie, DKSH, Janssen, Novartis, Pfizer, Salvatore Albani: None declared.