

Background: SjD is a chronic, heterogeneous autoimmune disease characterized by signs of oral and ocular dryness in patients who are either positive for anti-Ro/SSA and/or focus score positive on biopsies from labial/parotid salivary glands. Although involvement of salivary glands is a distinguishing disease feature, little is known about the transcriptomics of discrete cell populations in the glands.
Objectives: To determine the optimal dissociation approach and single-cell (sc) transcriptomics platform for the use of matched viably frozen and fixed minor salivary glands (MSG) biopsied from SjD patients and healthy controls (HCs); then, to expand the dataset using the optimal approach.
Methods: Viably frozen MSG (n=7; 2 SjD and 5 HCs) were thawed, dissociated, and counted, showing >90% viable cells. Samples were split and captured using 3’ and 5’ scRNA-seq, targeting to capture 8000-10000 cells. In addition, 25μm sections from formalin-fixed paraffin-embedded (FFPE) tissues containing 4-5 labial salivary glands (LSG) each were dissociated, counted, labeled following the 10X Flex/scFFPE protocol, and captured, yielding 8000-20000 cells per subject. Raw sequencing data were analyzed using 3’, 5’, or scFFPE analysis pipelines in CellRanger. Ambient RNAs were corrected (SoupX) and doublets detected (scDblfinder). Cells with feature counts <200 & >5000, mitochondrial percent >5%, and doublets were removed. Samples were merged, integrated, and batch corrected using Harmony (Seurat). Data were normalized, scaled, and dimensional reduction performed using UMAP (Seurat). Cell clusters were annotated using CellTypist and MSG reference data 1 . Cells were separated into immune and non-immune clusters and annotated using reference data from CellTypist or MSG reference data 1 . Differentially expressed (DE) genes in each cell type in SjD compared to HC were identified using pseudobulk approach (DESeq2). DE genes (p<0.05) were subjected to Ingenuity Pathway Analysis (IPA).
Results: Overall quality of the 3’ and 5’ approaches were similar. In comparison, scFFPE yielded similar feature/gene counts per cell (median = 5186 (3’), 7452 (5’), 4571 (scFFPE)), but was superior in the reduction of ambient RNA (or soup) and in identifying cell populations that were very low in 3’ and 5’ assays: seromucous acini, basal cells, macrophages, myoepithelial cells, among others. The scFFPE dataset was expanded to include 8 Ro+ SjD and 9 HCs, yielding 477,071 cells before QC, 371,454 after QC, among 23 cell states (Figure 1A). It revealed significantly DE genes in Ro+ SjD compared to HCs that mapped to common and different pathways between immune (Figure 1B and C) and glandular cells (Figure 1B and D). For example, many cell types showed dysregulation of Type I interferon, including downregulation (plasma cells, plasmacytoid DCs, naive B cells), upregulation (migratory DCs, DC1, serous and mucus acini, macrophages, mast cells), or no dysregulation (DC2, memory B, cytotoxic T, helper T, and natural killer cells).
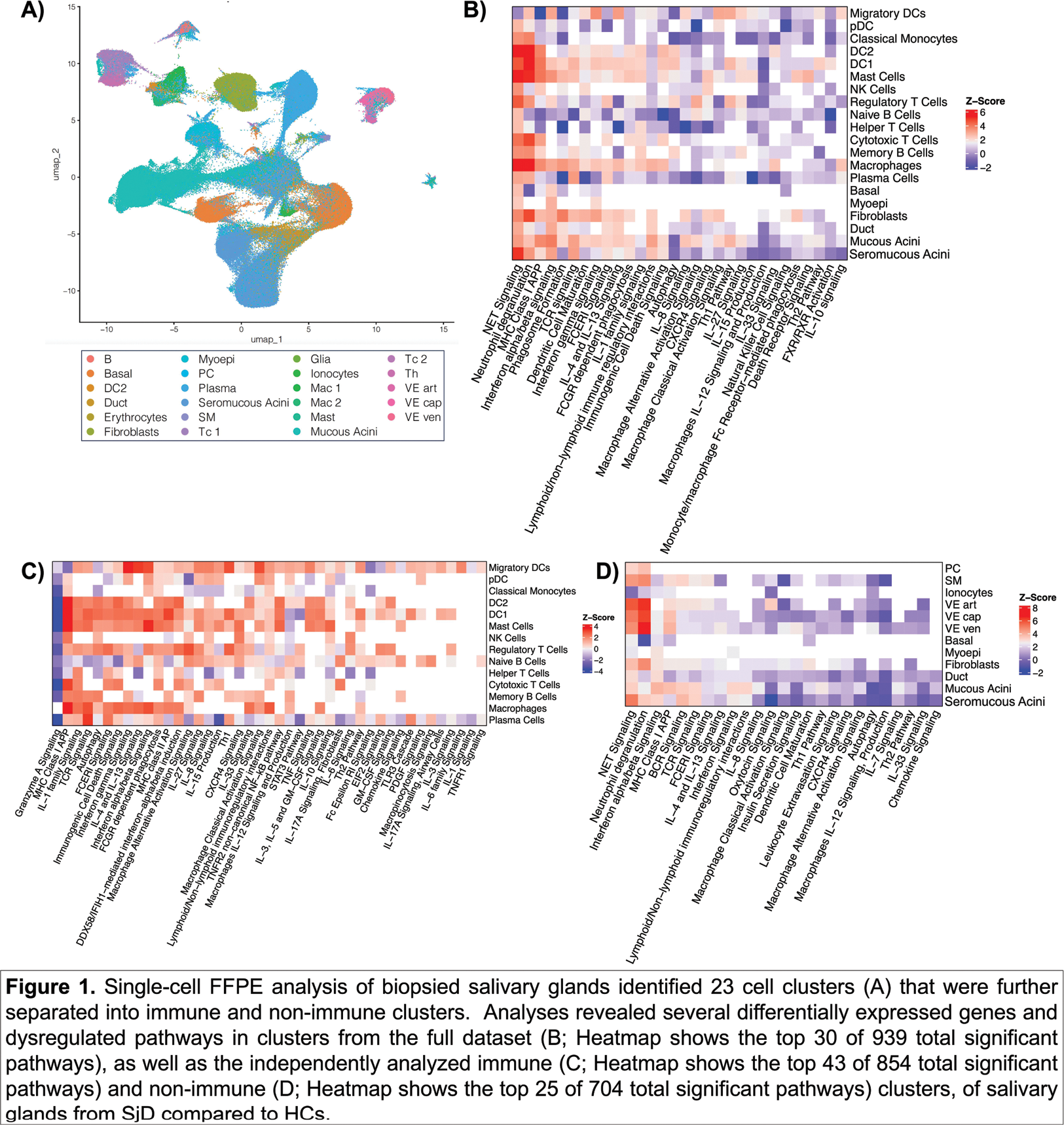
Conclusion: scFFPE yielded superior single-cell data compared to 3’ and 5’ using viable cells. Further, the data analysis method permitted discrimination of targeted/affected cell types from effector cell types. Expansion of the scFFPE pilot study showed many DE genes and dysregulated pathways. Ongoing work with AMP-AIM STAMP projects will further expand this dataset.
REFERENCES: [1] Huang N, et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat Med . 2021 May; 27(5):892-903.
Acknowledgements: National Institutes of Health: UM2 AR067678, UC2 AR081023, UC2 AR081032, UC2 AR081032-S1, UC2 AR081032-02S1, UC2 DE032254, UC2 AR081033, P30 AR073750, U54 GM104938, NIDCR 15-D-0051; Presbyterian Health Foundation; OMRF Institutional Funds; NIH, NCI Intramural Program; Jerome L. Greene Foundation.
Disclosure of Interests: Bhuwan Khatri: None declared, Anna M Stolarczyk: None declared, Matthew Caleb Marlin: None declared, Miles Smith: None declared, Cherilyn Pritchett Frazee: None declared, Margaret Beach: None declared, Eileen Pelayo: None declared, Zohreh Khavandgar: None declared, Paola Pérez: None declared, David E Kleiner: None declared, Stephen E Hewitt: None declared, Kevin Wei Received a sponsored research agreement from Gilead Sciences and 10X Genomics., Erin M Theisen: None declared, Kandice L Tessneer: None declared, Soumya Raychaudhuri: None declared, Michael B Brenner: None declared, Johann E. Gudjonsson: None declared, Nir Hacohen: None declared, Judith A. James: None declared, R Hal Scofield Received consulting fees from Johnson and Johnson Innovative Medicine (formerly Janssen) and Merk Pharmaceuticals., Stephen Shiboski: None declared, Astrid Rasmussen: None declared, Alan Baer Received consulting fees from Bristol Myers Squibb (BMS) and iCell Gene Therapeutics., A Darise Farris Grant/research support from Johnson and Johnson Innovative Medicine (formerly Janssen; ended 12/31/2023)., Caroline Shiboski: None declared, Blake M Warner Receives funding to support research from Pfizer, Inc., and Mitobridge, Inc., a subsidiary of Astellas Bio., Joel M Guthridge: None declared, Christopher J Lessard Grant/research support from Johnson and Johnson Innovative Medicine (formerly Janssen; ended 12/31/2023).