

Background: Haemophilus parainfluenzae ( H . para ) constitutes the majority of oral Haemophilus . Despite the wide presence of decreased oral abundance of Haemophilus or H . para in systemic or organ-specific autoimmune diseases, the significance of this reduction remains unexplored. Sjögren’s syndrome (SS) serves as a prototype of systemic autoimmune diseases. Salivary gland epithelial cells (SGECs) connect with the oral environment and coordinate the adaptive immune response, rendering them indispensable in the pathogenesis of SS [1]. This study focuses on the regulatory effects of H . para on SGECs and postulates an immunomodulation through the regulation of SGECs by H . para .
Objectives: To explore the significance of the reduced oral H. para in SS through its relevance to SGECs.
Methods: The salivary abundance of H. para in SS patients was determined through qPCR. A253 cells, derived from a human salivary gland with epithelial morphology, were pretreated by H. para and then co-cultured with CD4 or CD8 T cells. The proliferation of T cells was assessed using flow cytometry. To investigate the impact of H. para on A253 cells, RNA-sequencing was performed using the Illumina platform. Enrichment analysis was conducted by PANTHER against Gene Ontology (GO) terms, with FDR correction. TNFAIP3 expression was quantified using qRT-PCR and Western blot.
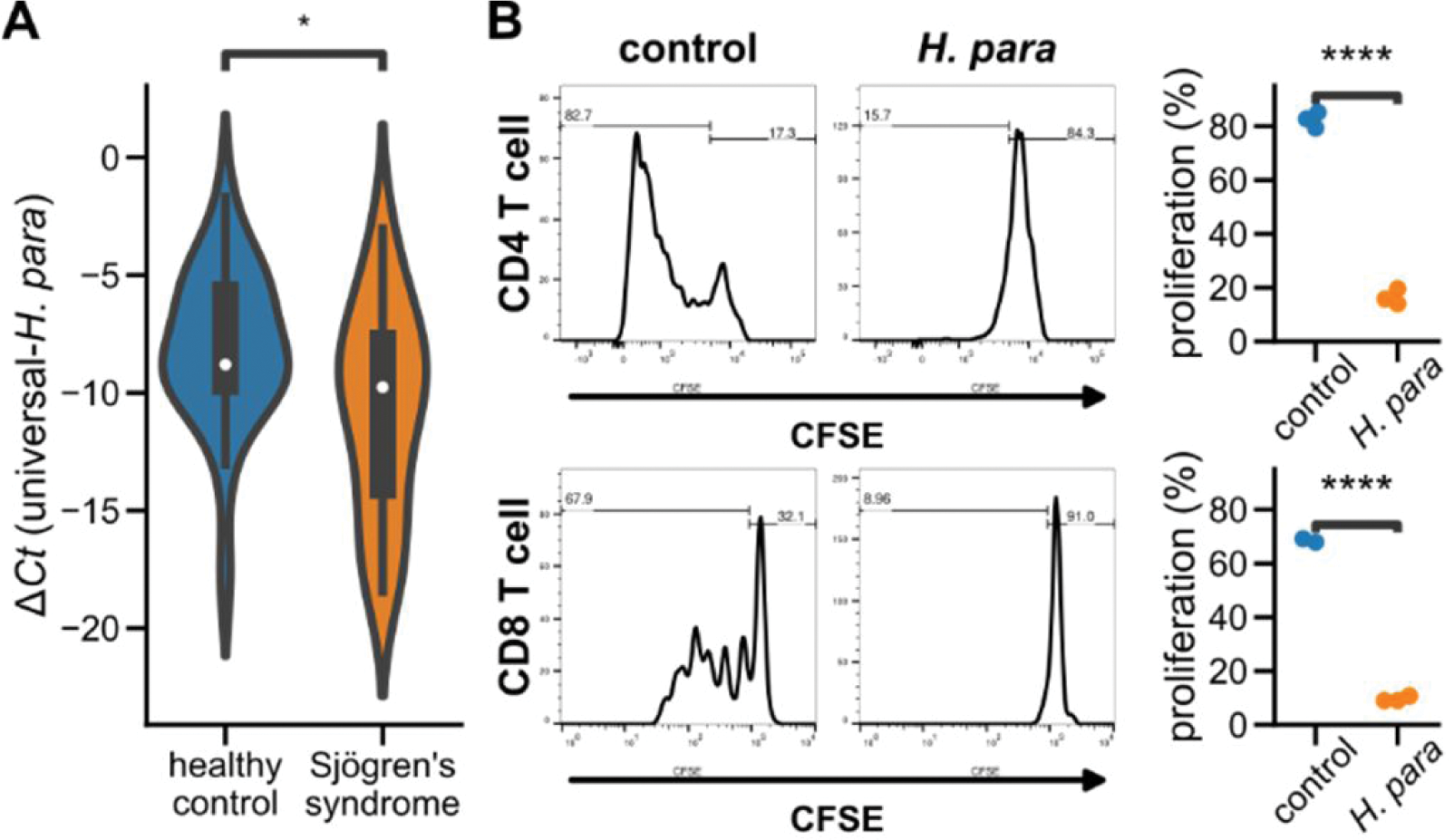
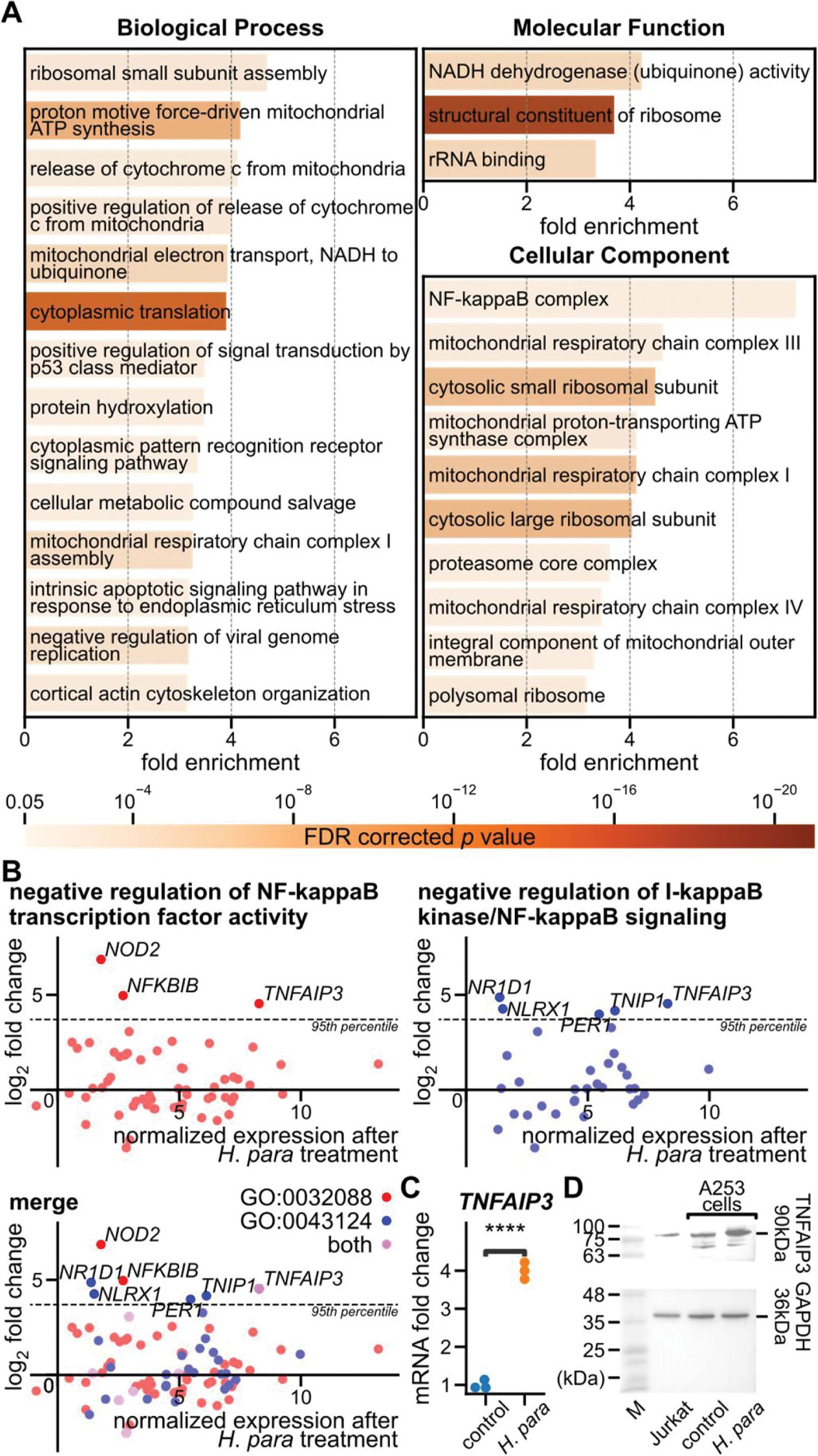
Results: Decreased H. para was found in saliva from 30 SS patients and 24 healthy controls (Figure 1A). H. para -pretreated A253 cells suppressed the proliferation of CD4 and CD8 T cells (Figure 1B). Transcriptomic analysis of H. para -pretreated A253 cells disclosed 7,291 upregulated and 7,709 downregulated genes. Enrichment analysis identified 27 GO terms (Figure 2A), including significant portions in translation and mitochondrial processes, showing active cellular response to stimuli and metabolic reprogramming. Notably, the GO term “NF-kappaB complex” exhibited the highest fold enrichment (7.22, FDR-corrected p = 0.024, Figure 2A), indicating a significant alteration in NF-κB complex. TNFAIP3 , ranking in the 97.5 th percentile of all upregulated genes, participated in the two GO terms involving the negative regulation of NF-κB (Figure 2B). The upregulation of TNFAIP3 was confirmed by qRT-PCR and Western blot (Figure 2C and D).
Conclusion: TNFAIP3 serves as a physiological negative-feedback mechanism following NF-κB activation. TNFAIP3 polymorphism was associated with SS [2], and the loss of TNFAIP3 in SGECs has been attributed to the pathogenesis of SS in vitro and in vivo [3,4]. We conclude that the reduction of oral H. para in SS may lead to inappropriate T cell activation, potentially through the loss of TNFAIP3 upregulation in SGECs. This study provides a provisional insight into dissecting the possible mechanisms of H. para in SS.
REFERENCES: [1] Verstappen, G. M., et al. (2021). Nat Rev Rheumatol , 17(6), 333–348.
[2] Khatri, B., et al. (2022). Nat Commun , 13(1), 4287.
[3] Sisto, M., et al. (2011). Histochem Cell Biol , 135(6), 615–625.
[4] X, W., et al. (2018). PLoS ONE , 13(8).
Salivary H. para in SS patients and the immunomodulatory effect of H. para through SGECs. A Violin plots depict the Δ Ct between H. para -specific and universal eubacteria primers. A more negative value indicates lower salivary relative abundance. * Mann-Whitney U test p < 0.05. B H. para -pretreated A253 cells were cocultured with T cells under anti-CD3/CD28 stimulation. Activated T cells underwent proliferation, leading to decreased CFSE intensity. **** Student’s t test p < 0.0001. CFSE: carboxyfluorescein succinimidyl ester.
Transcriptomic analysis and TNFAIP3 expression in H. para -pretreated A253 cells. A GO terms with the highest hierarchy, fold enrichment ≥ 3, and FDR-corrected p value < 0.05. B Gene expressions associated with the two GO terms related to the negative regulation of NF-κB. The y-axis denotes the log 2 fold change compared to control A253 cells. The horizontal dotted line indicates the 95 th percentile of the total upregulated genes. TNFAIP3 was the highest expressed gene after H. para treatment among the most upregulated genes in these two GO terms. C-D TNFAIP3 expression assayed by qRT-PCR and Western blot. **** Student’s t test p < 0.0001.
Acknowledgements: This study was subsidized by Ditmanson Medical Foundation Chia-Yi Christian Hospital Research Program (R109-001).
Disclosure of Interests: None declared.