

Background: The aim of this study was to investigate the LN transcriptome in depth to gain insight into underlying mechanisms and identify potential drug targets for LN.
Objectives: The aim of this study was to investigate the LN transcriptome in depth to gain insight into underlying mechanisms and identify potential drug targets for LN.
Methods: We analysed differentially expressed genes (DEGs) in peripheral blood from active LN (n=41) and active non-renal lupus (n=62) patients versus healthy controls (n=497) from the European PRECISESADS project (NTC02890121) and dysregulated gene modules in a discovery (n=26) and a replication (n=15) set of active LN cases. Replicated gene modules qualified for correlation analyses with serological markers, and regulatory network and druggability analysis.
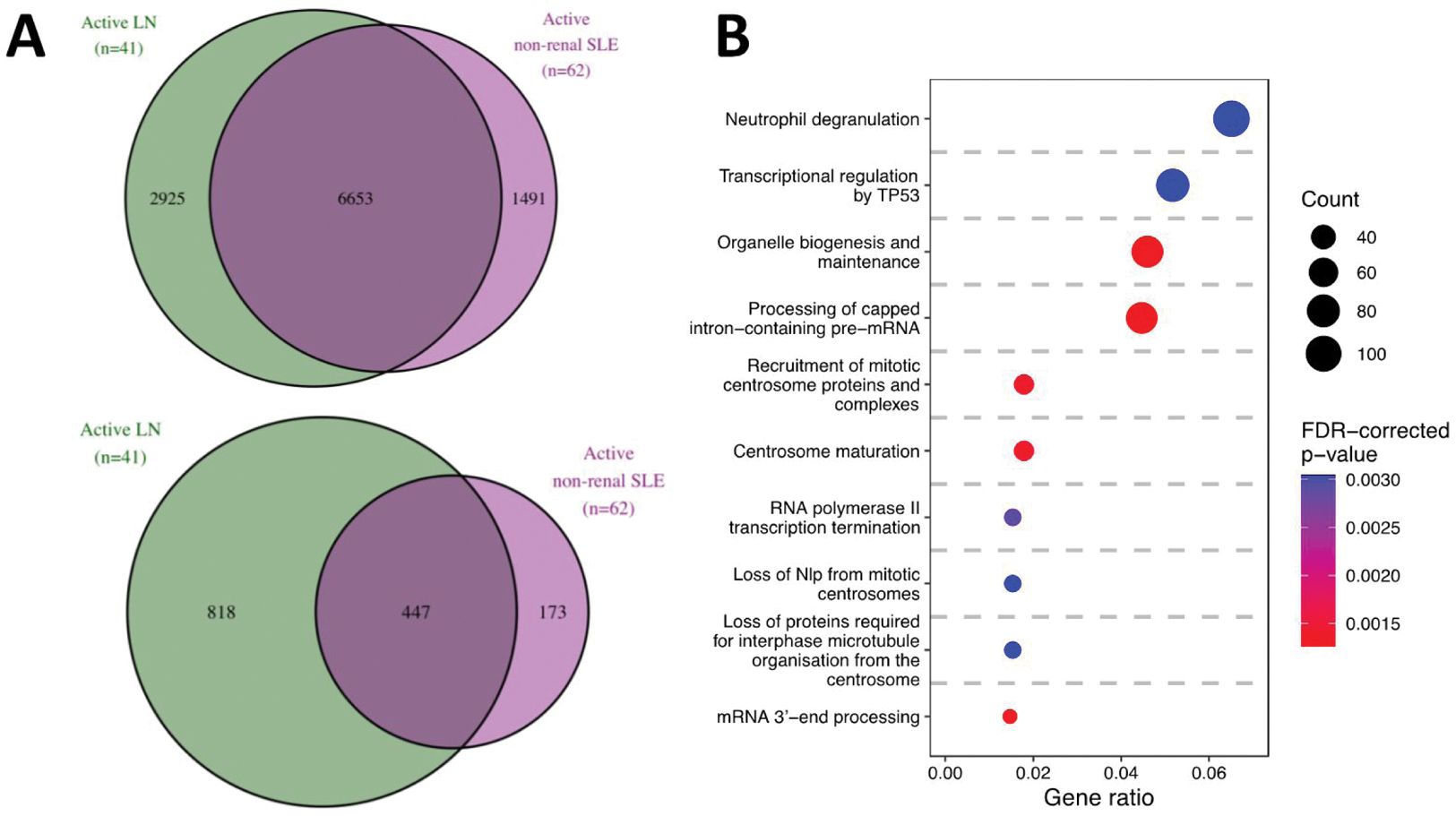
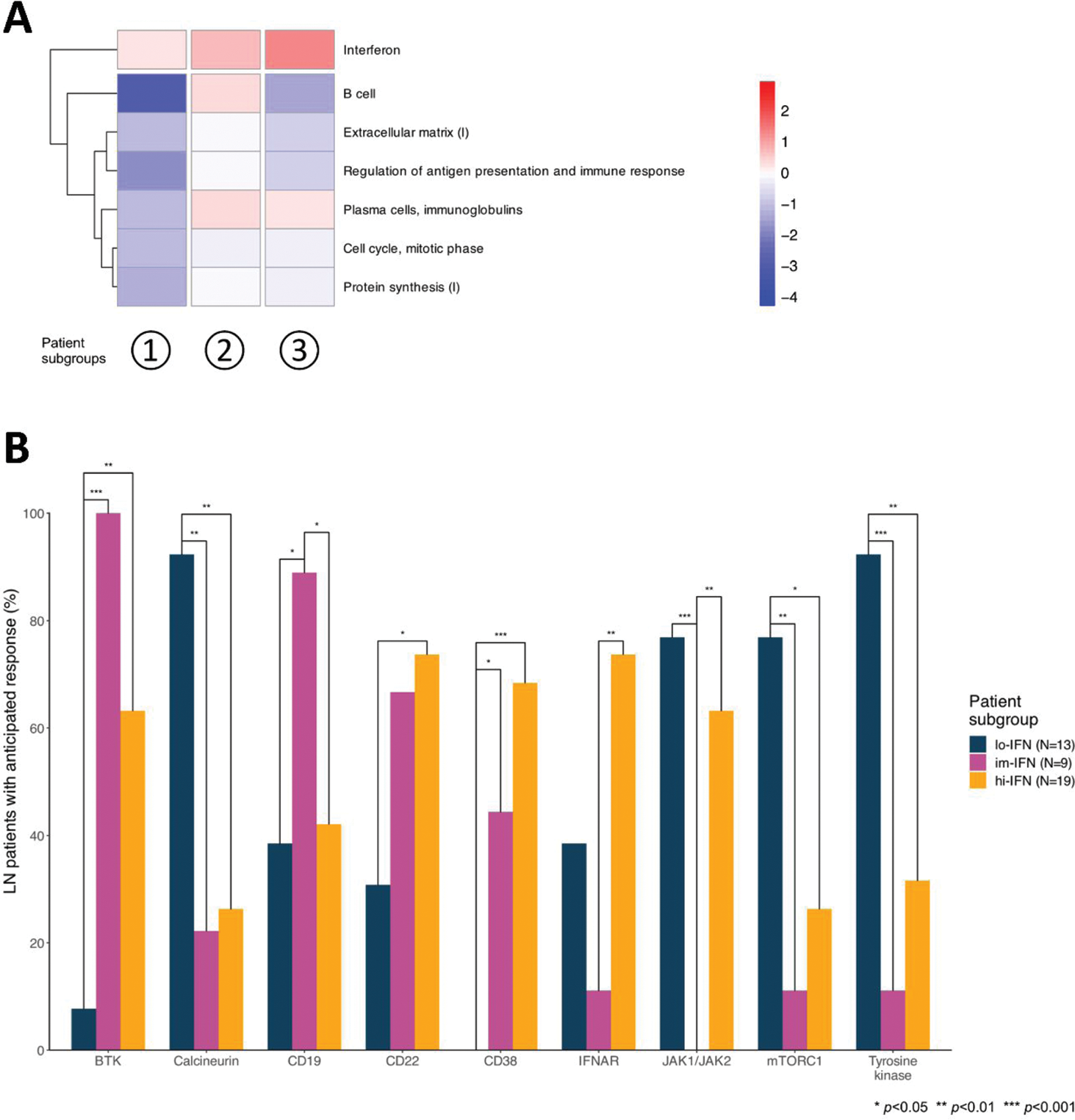
Results: We found 9578 DEGs in active LN vs HC and 8144 DEGs in active non-renal SLE vs HC. Active LN and active non-renal lupus patients shared 6653 DEGs, while 2925 DEGs were exclusively dysregulated in the LN group, denoting an LN-specific signature (Figure 1A). Among the latter, 818 DEGs displayed a |log 2 fold change (FC)| >0.58 (Figure 1A). The LN signature comprised DEGs associated with neutrophil degranulation, transcription regulation by TP53, and DNA damage response GO pathways (Figure 1B). Unsupervised co-expression network analysis revealed 20 dysregulated gene modules; seven showed prominent dysregulation in three distinct subgroups of LN patients (Figure 2A). These subgroups were classified based on the “interferon” (IFN) gene module upregulation into low, intermediate, and high IFN subgroups and showed differential dysregulation of the “B cell” and “plasma cells/immunoglobulins” modules. Drugs annotated to the IFN network included CC-motif chemokine receptor 1 (CCR1) inhibitors, programmed death-ligand 1 (PD-L1) inhibitors, and irinotecan, while the anti-CD38 daratumumab and proteasome inhibitor bortezomib showed potential for counteracting the transcriptomic signature associated with the “plasma cells/immunoglobulins” module. In silico analysis demonstrated that the low-IFN subgroup may benefit from calcineurin inhibitors while the intermediate-IFN subgroup may benefit from B cell targeted therapies (Figure 2B). High-IFN patients exhibited greater anticipated response to anifrolumab while the intermediate-IFN and high-IFN subgroups displayed greater anticipated response to daratumumab.
Conclusion: Interferon upregulation and B and plasma cell gene dysregulation patterns revealed three distinct LN patient subgroups, each demonstrating unique anticipated responses to existing or novel targeted therapies, paving the way for informed and tailored treatment strategies in LN. Of particular interest were irinotecan, CCR1 inhibitors, and PD-L1 antagonists interfering with IFN-mediated pathways, and bortezomib and daratumumab for their interference with plasma cells. While further studies are warranted to investigate the potential of these drugs, our findings contribute to a conceptual framework of precision medicine in the management of LN.
REFERENCES: NIL.
DEGs in patients with active LN and active non-renal SLE versus HC. (A) Venn diagrams showing DEGs (top) and the subset of DEGs that exceeded the |log 2 FC| >0.58 threshold (bottom) in patients with active LN vs HC (in green), and in patients with active non-renal SLE vs HC (in purple). (B) Reactome pathways from over-representation analysis, based on LN-specific DEGs in patients with active LN vs HC (panel A, n=2925).
Interferon (IFN) upregulation and B and plasma cell gene dysregulation patterns reveal three distinct lupus nephritis (LN) subgroups with differential anticipated response to targeted therapies. (A) Heatmap displaying the most discriminative gene modules, grouping the patients into low-IFN, intermediate-IFN, and high-IFN. Red and blue colours denote higher and lower gene dysregulation, as measured by z-scores, compared to healthy controls, respectively. (B) Bars depicting proportions of patients with an anticipated benefit from inhibition of selected drug targets across LN patient subgroups.
Acknowledgements: PRECISESADS Clinical Consortium.
Disclosure of Interests: Ioannis Parodis I have received research funding and/or honoraria from Amgen, AstraZeneca, Aurinia, Bristol Myers Squibb, Elli Lilly, Gilead, GlaxoSmithKline, Janssen, Novartis, Otsuka, and Roche., Julius Lindblom: None declared, Daniel Toro-Domínguez: None declared, Lorenzo Beretta: None declared, Maria Orietta Borghi: None declared, Jessica Castillo: None declared, Elena Carnero-Montoro: None declared, Yvonne Enman: None declared, Chandra Mohan: None declared, Marta Alarcon-Riquelme: None declared, Guillermo Barturen: None declared, Dionysis Nikolopoulos: None declared