

Background: One of the key players in autoimmune arthritis is T-cells. Both CD4 + and CD8 + T-cells have been implicated in mediating many aspects of synovial inflammation [1,2]. Oligoclonal populations of CD8 + T-cells specific for epitopes from Epstein-Barr virus, cytomegalovirus, and influenza virus were found enriched in the synovial fluid of patients with inflammatory arthritis [3]. There are also data suggesting that CD4 + T-cells from the inflamed synovium in rheumatoid arthritis (RA) represent activated Th1 cells that secrete IFNγ, which in turn orchestrates synovial inflammation [4–6]. Among CD4 + T-cells, regulatory T-cells (Treg), which are crucial in preserving immune homeostasis, are believed to be dysfunctional in autoimmunity. In the inflamed joints of juvenile idiopathic arthritis (JIA) patients, a higher frequency of clonally expanded Tregs was observed than conventional CD4 + T-cells [7].
Mapping the immune phenotypes in synovial fluid of juvenile and adult patients with chronic autoimmune arthritis.
Decoding the synovial fluid T-cell receptor repertoire across different arthritis groups and T-cell subsets.
Identifying T-cell receptors specific to HLA-B27 associated spondyloarthropathies.
Methods: 27 patients were recruited: 6 had oligoarticular JIA (oJIA), 4 had enthesitis-related JIA (JIA-ERA), 3 had juvenile psoriatic arthritis (JIA-PsA), 5 had adult psoriatic arthritis (PsA), 5 had RA, 3 had Lyme arthritis (borrelia), and 1 had spondyloarthritis (SpA). Their synovial fluid was collected and processed into mononuclear cells (SFMC). Single-cell RNA-seq and TCR-seq were performed on the SFMC. Bulk TCR-seq was done on the CD4 + , CD8 + , and Treg fractions.
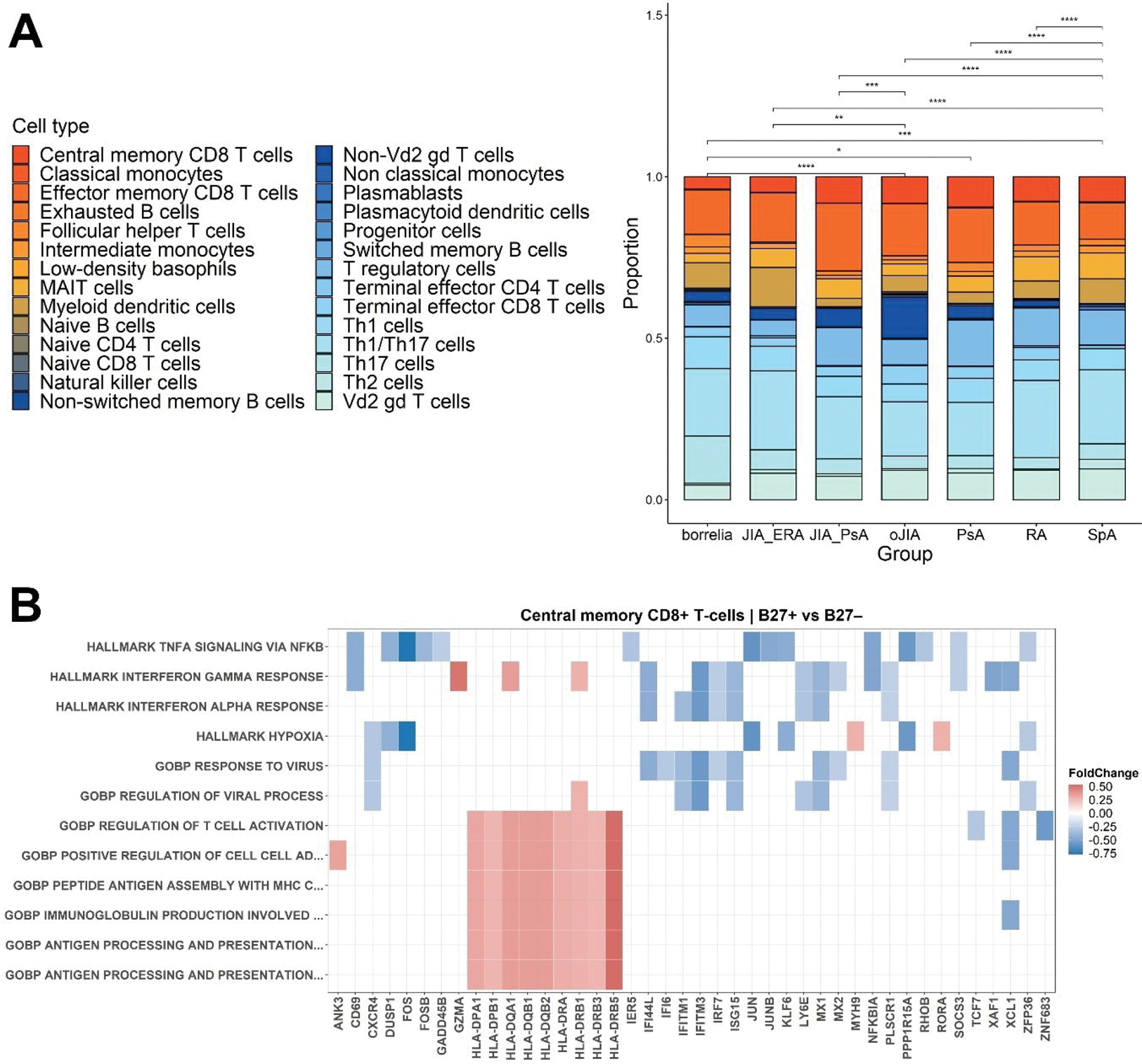
Results: Our single-cell sequencing data indicate that among the 28 identified cell phenotypes, Th1/Th17 cells were the dominant population among all patient groups, having the highest proportion out of the total T-cell count in JIA-ERA (Figure 1A). Furthermore, compared to B27 – patients, B27 + patients had upregulated HLA-DP, HLA-DQ, HLA-DR, and GZMA genes in central memory CD8 + T-cells, implying an increase in T-cell activation and cytotoxic activity (Figure 1B). Gene set enrichment analysis also suggests pro-inflammatory pathways (e.g., TNFα, IFNα, and IFNγ signalling), and T-cell activation in response to viruses in B27 + patients.
Single-cell sequencing data of 27 patients having arthritis. (A) Stacked bar plot of the identified cell types, statistical significance was analysed by Wilcoxon test with Bonferroni correction (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001); (B) Differential gene expression and gene set enrichment comparison of central memory CD8 + T-cells between B27 + and B27 – patients.
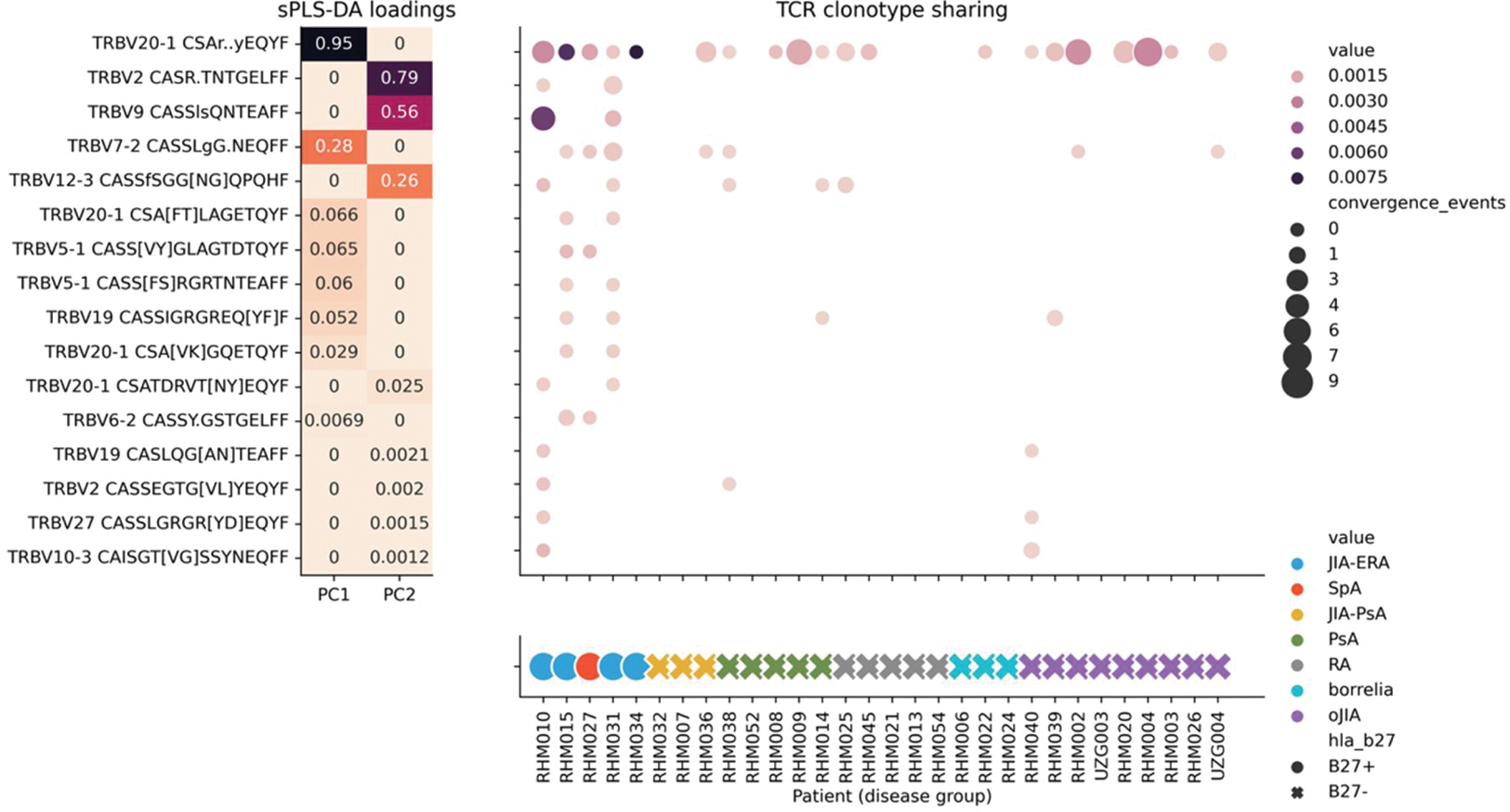
From the bulk TCRβ sequencing data (Figure 2), we observed that HLA-B27 + patients shared broad TCRBV20 clusters specific for various viral epitopes, indicating a possible cross-reactive response. Furthermore, this method highlighted TCR clusters with sequence similarity to those known to be associated with HLA-B27 pathologies, as well as TCRs with unknown specificity, but a conserved GRGR motiflet.
Integrative comparison of TCRβ clusters between HLA-B27 + and HLA-B27 – patients. TCR sequences were clustered using ClusTCR and sparse supervised dimensionality reduction was applied to retain the eight most informative TCR clusters per principal component.
Conclusion: The findings in this study provide insights into how TCR repertoires differ between arthritis groups, patients’ ages, T-cell subsets, and whether viral/bacterial infection contributes to arthritis onset and flares. Such knowledge will greatly contribute to the understanding of T-cells’ and TCRs’ participation in the underlying pathophysiology of arthritis.
REFERENCES: [1] Black, A.P.B. et al. Arthritis Res Ther 2002 , 4 , 177.
[2] Skapenko, A. et al. Arthritis Res Ther 2005 , 7 , S4.
[3] Fazou, C. et al. Arthritis and Rheumatology 2001 , 44 , 2038–2045.
[4] Cañete, J.D. et al. Ann Rheum Dis 2000 , 59 , 263–268.
[5] Davis, L.S. et al. Arthritis Res Ther 2000 , 3 , 54.
[6] Möller, B. et al. Rheumatology 2001 , 40 , 302–309.
[7] Mijnheer, G. et al. Elife 2023 , 12 , e79016.
Acknowledgements: NIL.
Disclosure of Interests: None declared.