

Background: Despite advancements in the treatment of rheumatoid arthritis (RA), optimal disease control remains a challenge in the difficult-to-treat RA (D2T) subset of patients, often resulting in persistent symptoms, especially pain. The etiology of pain in D2T RA is multifactorial, involving factors such as ongoing inflammation, altered pain processing, structural damage, and various physical and psycho-social comorbidities. Recent literature suggests that peripheral blood mononuclear cells (PBMC) can reflect the pathophysiological mechanisms of the central nervous system.
Objectives: Our main goal was to compare the transcriptomic profiles of PBMCs of RA patients with a special focus on D2T RA and healthy controls (HC) to identify characteristic differences in molecules and signaling pathways and to reveal the connection between pain and inflammation.
Methods: The research was carried out in two centers, including 31 HCs and 48 RA patients, out of whom 30 were classified as D2T RA. Based on pain and inflammatory parameters, patients were divided into different subgroups: a) high inflammation (C-reactive protein (CRP)>5 mg/l or erythrocyte sedimentation rate (ESR) > 25 mm/hr) and high pain (Pain Visual Analogue Scale(VAS)>50), b) high inflammation and low pain (PainVAS≤50), c) low inflammation (CRP≤ 5 mg/L or ESR≤25mm/hr) and high pain and c ) low inflammation and low pain . Transcriptomic analysis was performed on the total RNA isolated from PBMCs by next-generation sequencing, and the results were evaluated using bioinformatics tools (Figure 1).
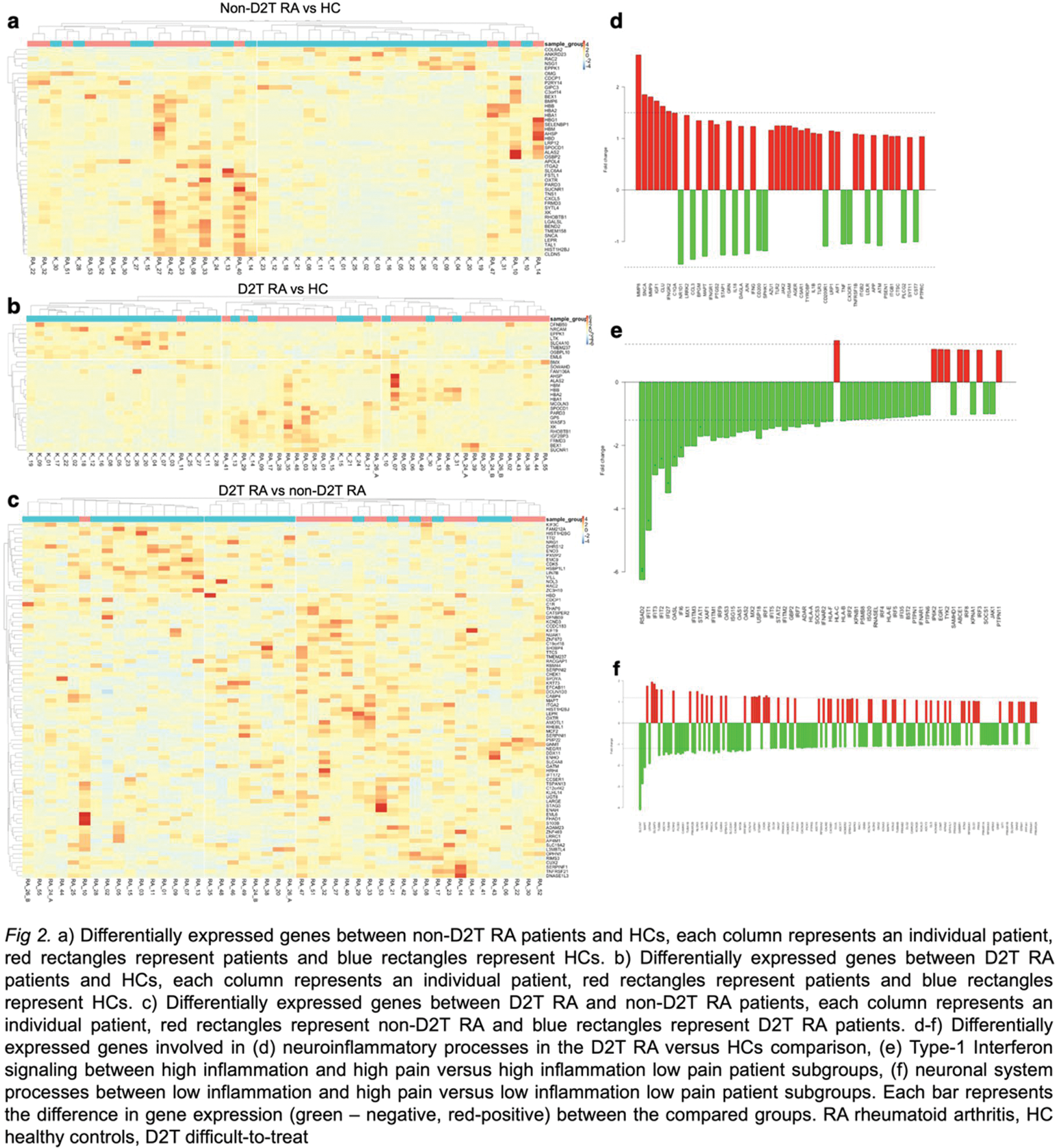
Results: In the non-D2T RA compared to HCs, 8 upregulated and 40 downregulated genes were determined mainly involved in inflammatory mechanisms such as leukocyte activation and migration ( e.g., ITGA1 encoding the alpha 1 subunit of integrin receptors ) and regulation of immune responses (e.g., CXCL5 encoding the CXC motif chemokine ligand 5 ) (Figure 2a). When comparing D2T RA patients to HCs, 8 downregulated and 20 upregulated genes were found. However, in this case, in addition to inflammatory processes, many genes (e.g., SCNA encoding a protein regulating synaptic vesicle trafficking and MMP8 and 9 encoding matrix metalloproteinases ) could be linked to neuroinflammatory responses, microglial activation, and sensory pain perception too (Figure 2b and d). The comparison between D2T RA and non-D2T RA patients identified 16 upregulated and 66 downregulated genes, from which many could be linked to neuronal differentiation functions and altered synaptic signaling (e.g., NEGR1, neuronal growth regulator 1, S100B encoding a calcium-binding protein, etc.) (Figure 2c). Based on pain and inflammatory parameters, different subgroups of RA patients were compared. In patients with high inflammation and high pain, the vast majority of the altered genes involved in Type I interferon signaling and regulation (e.g., RSAD2, IFIT1, IFIT2, etc.) was found to be downregulated compared to patients with high inflammation and low pain. (Figure 2e ) Patients with low inflammation but having high pain compared to low inflammation but low pain showed significant alterations in the expression of genes involved in several processes related to inflammation and the neuronal system (e.g., SLC1A7, MAPT, etc.). (Figure 2f).
Conclusion: Our study reveals specific transcriptomic variations between RA patients and HCs, as well as characteristic differences between non-D2T RA versus D2T RA patients, mainly involving neuronal activation and sensitization processes. In the subgroups of RA patients with different pain and inflammatory parameters, the downregulation of Type-1 Interferon signaling seems to contribute to the pain intensity of patients with high inflammation, whereas alterations of neuronal system processes seem to play an important role in pain accompanying low inflammatory activity. Therefore, PBMC transcriptomics appears to be useful for identifying differentially expressed genes associated with pathophysiological mechanisms involved in inflammatory processes and pain sensitization. Confirming hypotheses generated with this unbiased omics approach can facilitate the development of novel therapies.
REFERENCES: NIL.
Acknowledgements: Funding: HUN-REN–PTE Chronic Pain Research Group (14017), OTKA Grant (K 138046) and (K 131479), TKP2021-EGA-29, the National Brain Research Program and the Richter Gedeon Talentum Foundation with the help of „Richter Gedeon PhD Scholarship” for LGT. Figure 1. was created with biorender.com.
Disclosure of Interests: None declared.