

Background: Systemic autoinflammatory disorders (SAIDs) include a group of diseases, which are associated with genetic variations resulting in dysregulation of innate immunity and hyperinflammatory response. Pathogenic and likely pathogenic variants in certain genes have been linked to an already defined phenotype, but screening of more patients with some autoinflammatory features, but no diagnosis, has usually revealed several variants of uncertain significance (VUS) and/or variants in more than one autoinflammatory gene.
Objectives: We herein investigated the relationship between phenotypes and genotypes of the patients who were screened for autoinflammatory genes and identified as carriers of variants in ≥2 genes in a tertiary referral center.
Methods: Charts of the patients who had been referred to our center with a potential diagnosis of an autoinflammatory disorder between April 2019 and January 2023, and the results of their genetic analyses were evaluated. All patients had the results of genetic screening of at least 22 genes, which were associated with different autoinflammatory disorders, and analyses covered the exons and 10 base pair intron borders of the selected genes and were carried out by using the Ion Torrent platform or Illumina platform.
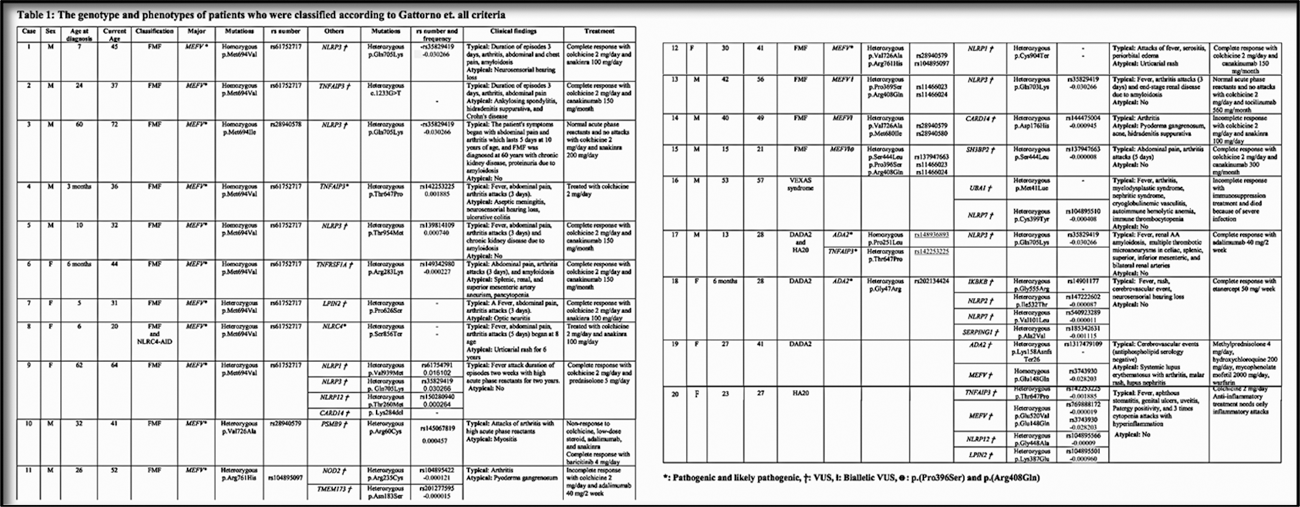
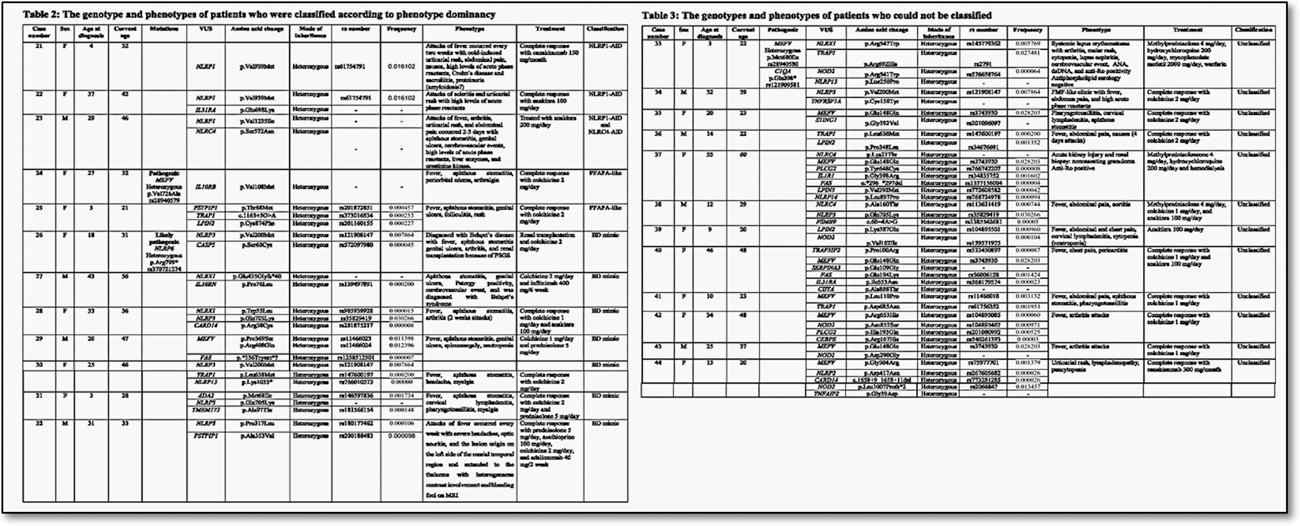
Results: We evaluated 146 referred patients (60 male, 85 female, mean age 41.0±12.6, range 19-82), and 44 of them (30.1%) were identified as having ≥2 gene variants. By using the recent classification criteria for autoinflammatory recurrent fevers, fifteen out of 44 (34.0%) patients were classified as FMF. Among the patients with the pathogenic MEFV mutations, variants of the additional genes may have had an effect on the phenotype such as sensorineural hearing loss in one with NLRP3 VUS. One patient with pathogenic MEFV and NLRC 4 mutations was classified as FMF and NLRC4-AID. Since the Gattorno et al. criteria were aimed at only 4 common SAIDs, 5 (11.3%) additional patients could be diagnosed with other disorders such as VEXAS syndrome (n=1), deficiency of ADA2 (DADA2, n=2), A20 haploinsufficiency (HA20, n=1), DADA2 and HA20 combination (n=1) (Table 1). The remaining patients were grouped as having NLRP1-AID (n=2), NLRP1-AID and NLRC4-AID (n=1) combination, PFAPA-like (n=2) and Behçet disease-mimic (n=7) according to their dominant phenotype (Table 2). Twelve (27.2%) patients could not be classified into any groups (Table 3). Two of these cases had pathogenic, likely pathogenic variants, or VUS, and the remaining had between 2-7 different VUS combinations. NLRP1 variants were identified in 5 (11.3%) cases and the most common clinical finding was recurrent urticaria (n=4). All of the NLRP1 gene variants were classified as VUS; one of them (p.Val939Met) was a relatively common variant (allele frequency, 0.01), one of them (p.Val1235Ile) was not reported before, and the other one (p.Cys904Ter) was expected to have a pathogenic role due to early termination of the protein. Case 9 had an atypical FMF phenotype with a pathogenic variant in the MEFV and VUS in others, including the NLRP1, NLRP3, and NLRP12.
Conclusion: The criteria set developed by Gattorno et al. have some limitations in practice since they cover only CAPS, FMF, TRAPS, and MKD, and are not validated in adults. Although those patients with pathogenic variants could be classified easily by using the criteria sets, the contribution of additional variants to their phenotypes needs further investigation. Four patients with recurrent urticaria as the most common clinical feature in association with 3 different VUS in the NLRP1 gene may suggest adding this gene to the list of autoinflammatory variants associated with urticarial manifestations, which may expand the spectrum of NLRP1-AID. An important group of adult patients may have an unclassified systemic autoinflammatory disorder, mainly associated with a combination of VUS in different genes, and this “mixed autoinflammatory disorder (MAID)” phenotype needs to be followed carefully for the evaluation of long-term prognosis.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.