

Background: Systemic lupus erythematosus (SLE), a clinically diverse autoimmune disease, poses challenges in predicting its varied manifestations. While soluble mediator profiles have been used, their predictive accuracy is still limited.
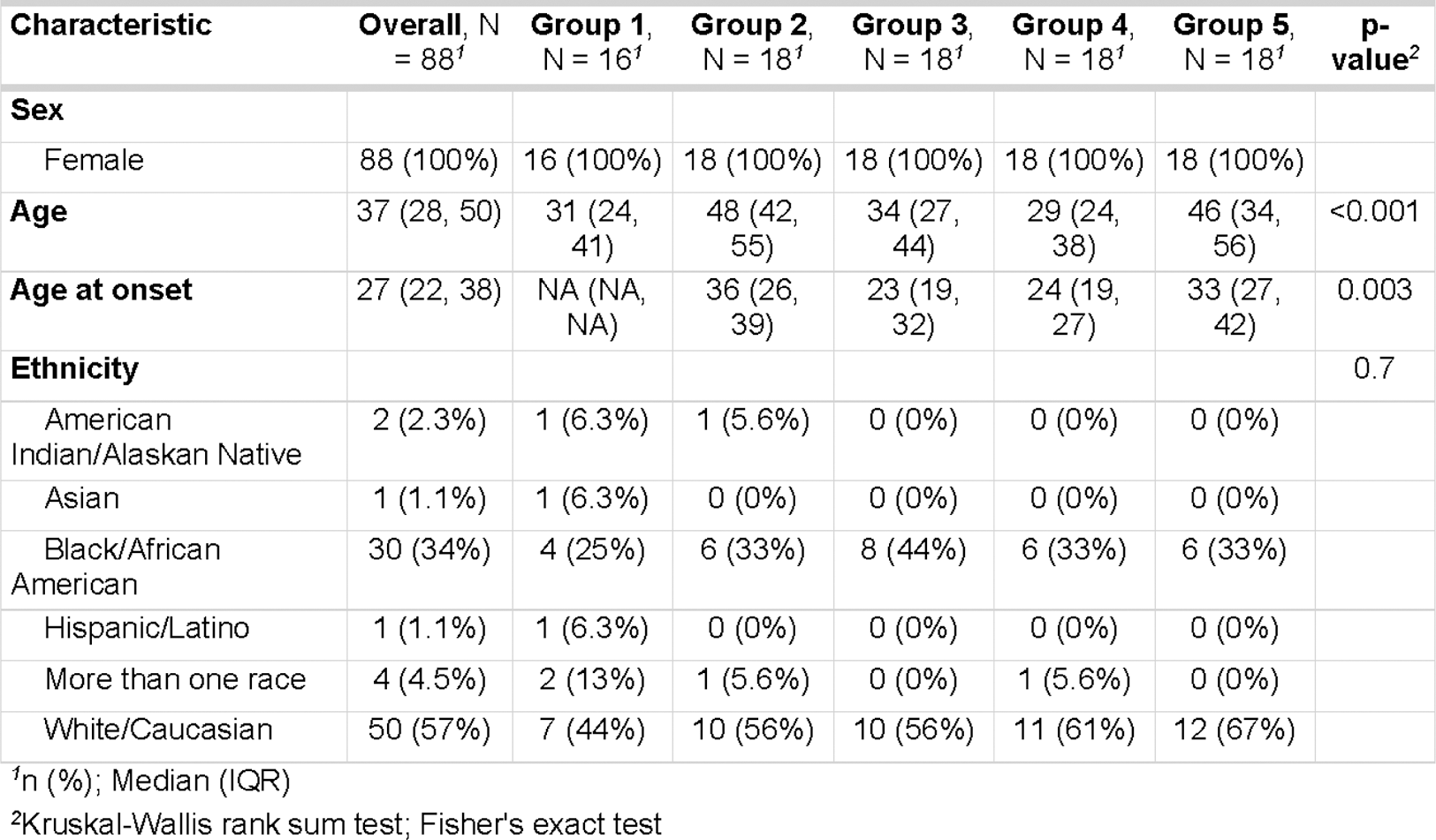
Objectives: This study aimed to evaluate the potential of soluble human proteomics and autoreactomics in predicting severe SLE manifestations. We included 88 plasmas from 16 healthy controls and 72 SLE patients categorized into groups based on clinical characteristics: Group2 - without renal disease nor anti-nuclear antibodies, Group3 - with anti-dsDNA but no renal disease, Group4 - both anti-dsDNA and renal disease, and Group5 - strong positive anti-RNA autoantibodies with some renal disease.
Methods: Here, we utilized a high-throughput, sensitive, and highly multiplexed human proteomic assay (Olink Explorer NGS) consisting of 1472 analytes as well as their autoantibody profiles.
Results: Univariate analysis showed that there are 186 dysregulated proteins in Group2, 398 proteins in Group3, 444 proteins in Group4, and 375 proteins in Group5 compared to controls. Pathway analysis revealed distinct upregulation in toll-like receptor, interleukin-10 response, and cytokine/chemokine signaling in Group3, Group4, and Group5. Notably, interferon-gamma production/signaling was prominent only in group 4 with renal disease. Unsupervised dimensional reduction analysis using UMAP identified four clusters, with cluster 4 comprising patients with renal disease. Cluster 4 showed significantly higher levels of HAVCR2, BST2, IL10, CD300, and IFNG than other clusters. Finally, an iterative feature-selection Random Forest machine-learning model based on HAVCR2, BST2, TNFRSF1B, CXCL9, CD300, and IFNGR was able to predict patients with renal involvement with ~91.0% accuracy. Of interest, HAVCR2 and BST2 are both highly expressed in glomeruli and renal tubules, suggesting the tissue-specific damaging signatures with inflammatory soluble mediators may provide superior manifestation prediction potential.
Conclusion: The study underscores the importance of a comprehensive approach, combining molecular signatures and inflammatory mediators, for enhanced prediction and monitoring of SLE severity.
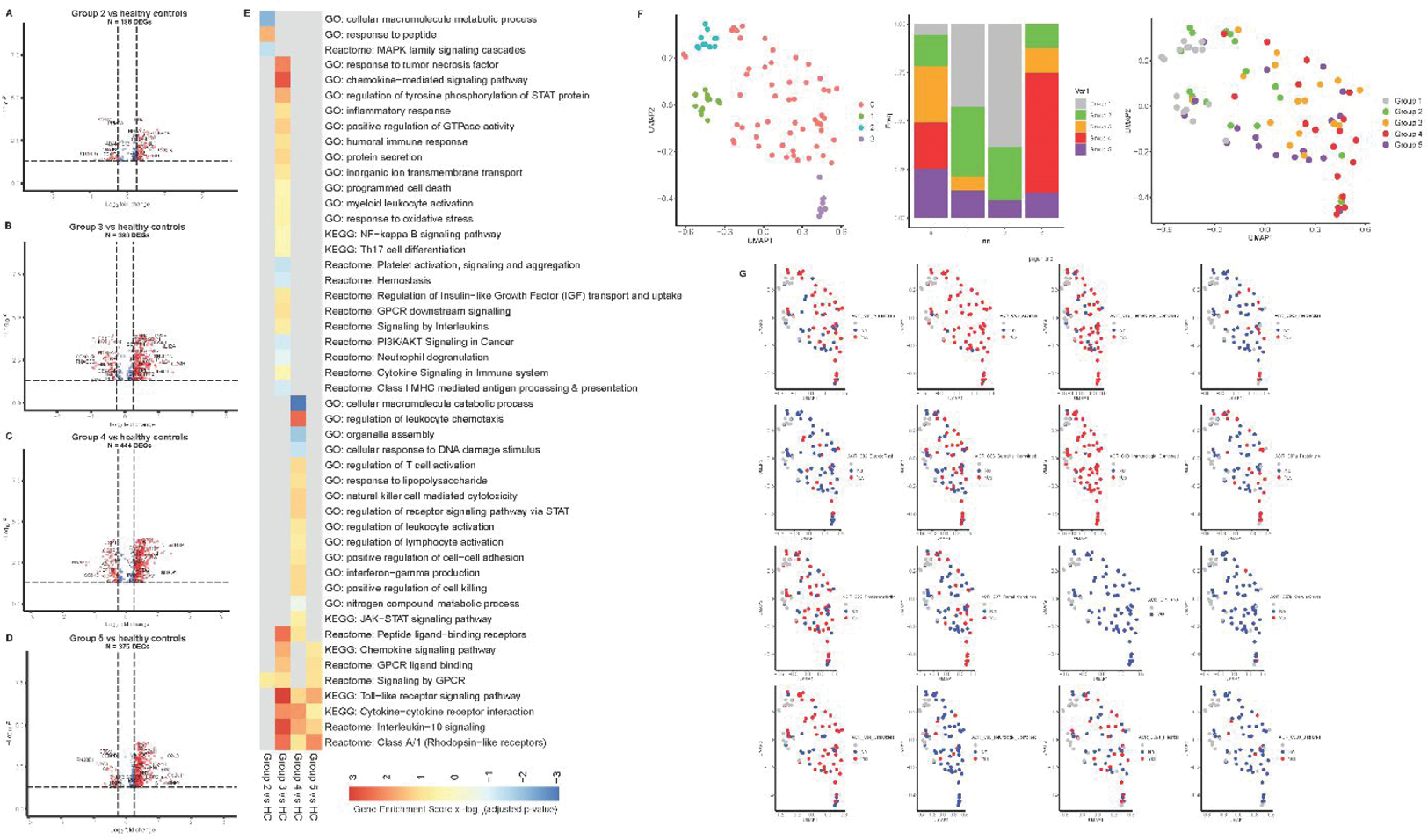
Table 1.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.