

Background: ANA-RMDs arise from a common At-Risk population defined by ANA antibodies with or without clinical symptoms. Accumulating evidence indicates that ANA positivity is a complex and dynamic immune dysregulation in which the majority of individuals remain clinically well. Most previous analyses of this population have focussed on gene expression or antibody status, but it is unclear how these are regulated at a functional protein level.
Objectives: To investigate immune dysregulation using the functional protein expression and identify biomarkers for the progression to SLE in ANA+ at-risk populations.
Methods: We selected 103 ANA At-Risk subjects for proteomics analysis from a subgroup of two Leeds prospective Cohort databases (CONVAS and DEFINITION). The eligibility criteria required individuals with IIF ANA-positivity (≥ 1:80) with or without specific autoantibody positivity from multiplex immune assay, not fulfilling SLE or other RMD classification criteria. Stored serum was analysed by Proximity Extension Assay (PEA) technology (Olink®, Uppsala, Sweden) using the EXPLORE Inflammation I and II panel, comprising 764 proteins. Progressors were subjects who met SLE or pSS criteria at follow-up (n=36). Samples from healthy controls (HC, n=10) and SLE (n=32) were also analysed. Protein quantification was reported in-log10 Normalized Protein eXpression (NPX) (adjusted p-value) relative protein quantification unit on a log2 scale and analysed using the Olink-analyse package in R.
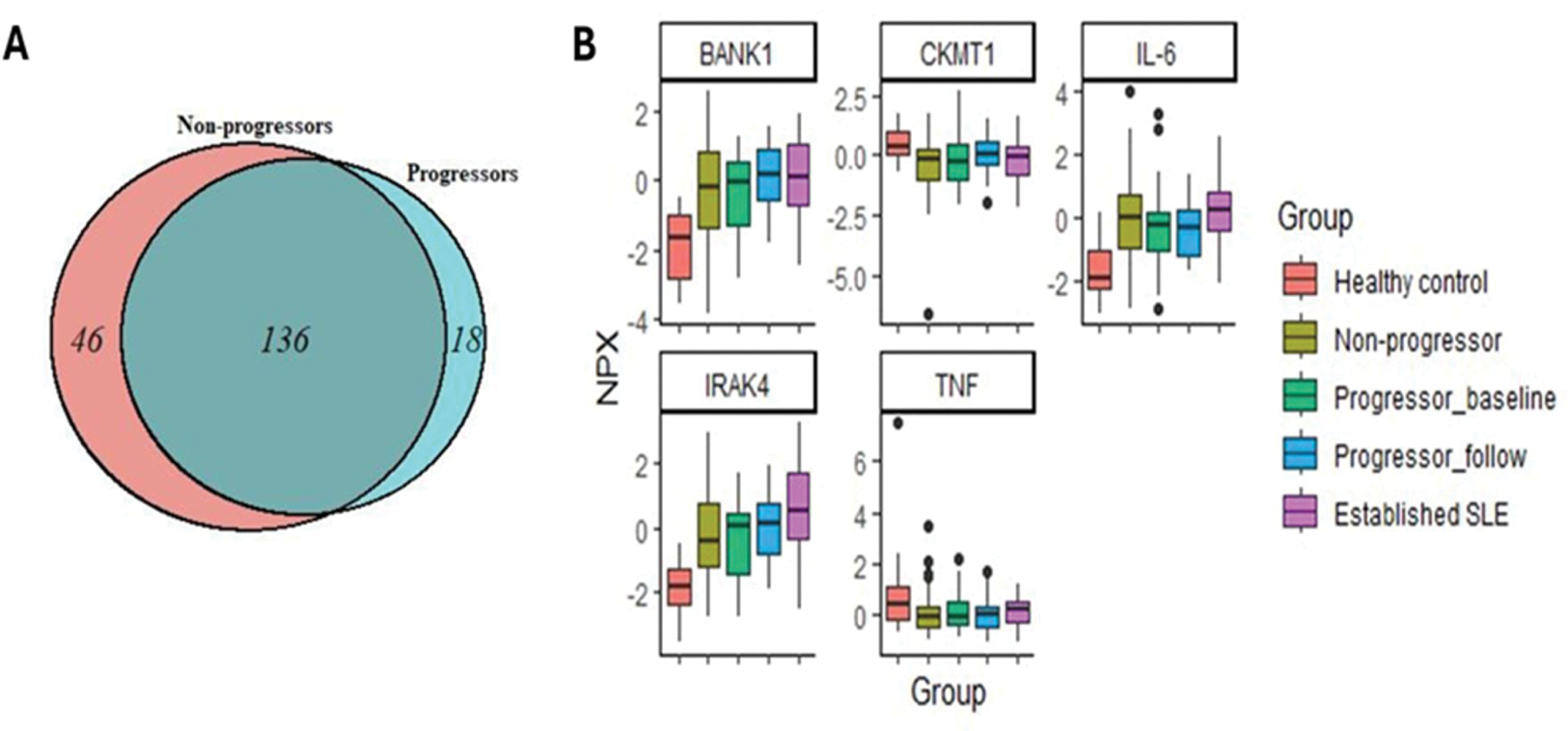
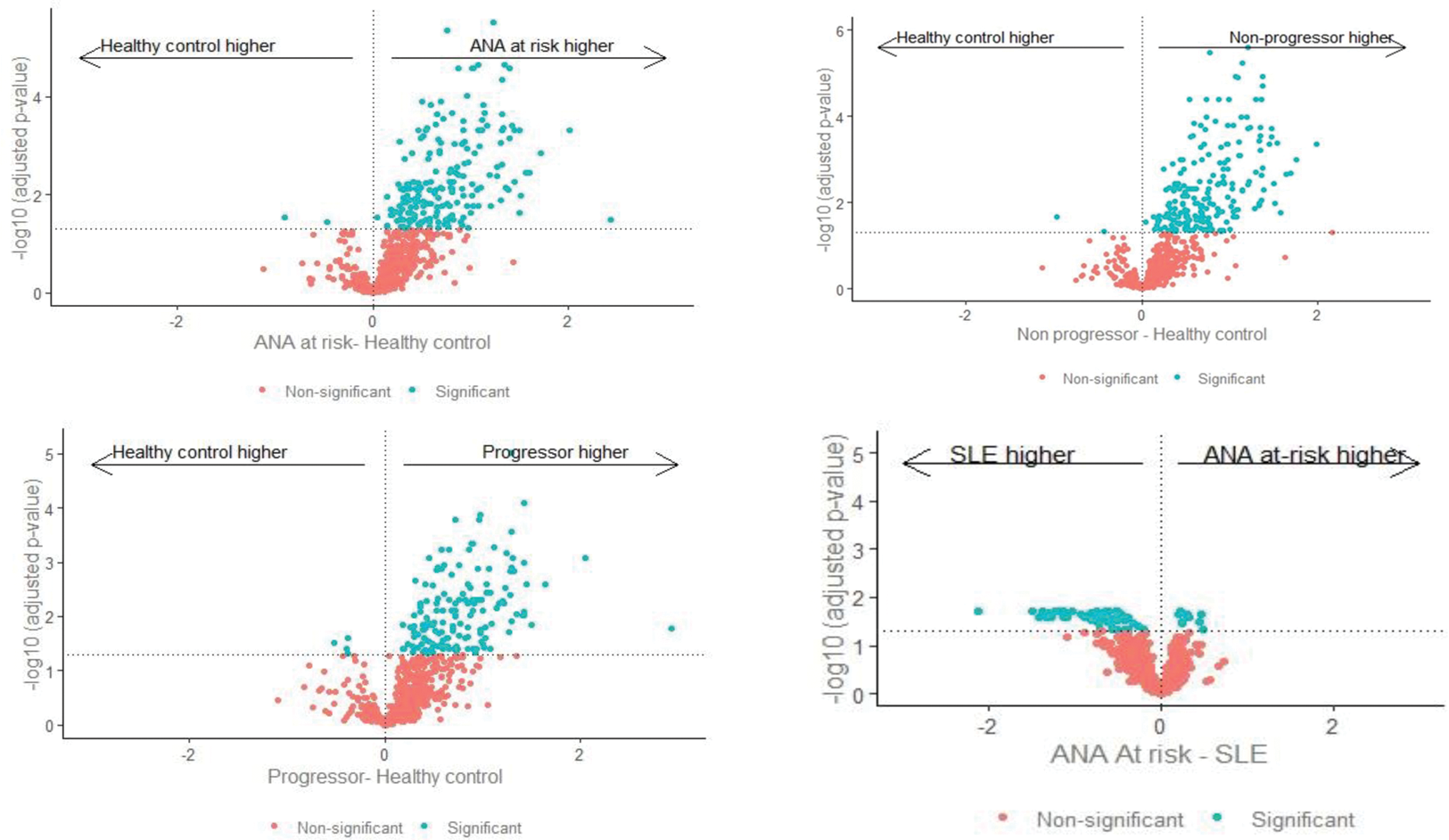
Results: Overall, 200 proteins significantly differed (adjusted p-value <0.05, Figure 1A) between HC and At-Risk individuals. Of these, 17 were significantly higher in progressors only, 45 were significantly higher in non-progressors only, and 136 were significantly in both progressors and non-progressors. Two proteins were significantly lower in At-Risk individuals than HC (Figure 2). However, there was no difference in protein levels comparing progressors and non-progressors. In the At-Risk population, significantly increased proteins with well-characterised immune functions included IFN-I-related proteins (e.g. CXCL9), B cell-related proteins (e.g. B2M, BAFF, BANK1, BCR), IFN-gamma and lambda, IRAK 1 and 4, IL6, IL12B, IL1B. TNF and CKMT1A were significantly lower in At Risk individuals compared to HC (Figure 1B). Proteins uniquely increased in progressors included CXCL10 and CXCL17, OX40, IL32, IL12B, C3, TACI, BCMA. Proteins uniquely increased in non-progressors included CXCL14, CXCL17, IL18. Few differences were found between At-Risk and SLE patients.
Conclusion: This study demonstrated a marked dysregulation of functional protein profile among ANA-positive individuals even in the absence of clinical autoimmunity. This adds to the understanding of ANA positivity as a complex and dynamic equilibrium in which SLE is not inevitable. Future work will validate key findings as clinically applicable biomarkers.
REFERENCES: NIL.
(A) Venn diagram showing the significant quantified proteins between ANA-positive at-risk and HC. (B) Boxplot demonstrates the significantly quantified protein with well-characterised immune functions between the groups
Volcano plot demonstrated significant protein quantification in each ANA+ at-risk subgroup compared with HC and SLE.
Acknowledgements: NIL.
Disclosure of Interests: Porntip Intapiboon Novatis, Amgen, Lucy M. Carter Alumis, Jack Arnold Alumis, Khaled Mahmoud: None declared, Md Yuzaiful Md Yusof Roche,Novartis and Alumis, Aurinia Pharmaceuticals and UCB, Edward M. Vital Roche, GSK, AstraZeneca, Aurinia Pharmaceuticals, Lilly and Novartis, Roche, AstraZeneca and Sandoz.