

Background: Systemic lupus erythematosus (SLE) is a prototypic systemic autoimmune disease. Despite advancements in treatment, the patients still suffer from flares. Since flares impact both prognosis and organ damage, the prevention of flares is a key therapeutic goal. However, gene expression profiles associated with flares remain poorly understood, making it challenging to implement individualized treatment strategies.
Objectives: To identify gene expression profiles predicting subsequent flares in patients achieving lupus low disease activity state (LLDAS).
Methods: We conducted a prospective cohort study. We set up two cohorts by extracting SLE patients achieving LLDAS from our two bulk RNA-seq studies, ImmuNexUT[1,2] and AMED-SLE[3], designating a test cohort and a validation cohort, respectively. We followed up these cohorts, assessing flares based on SELENA-SLEDAI flare index.
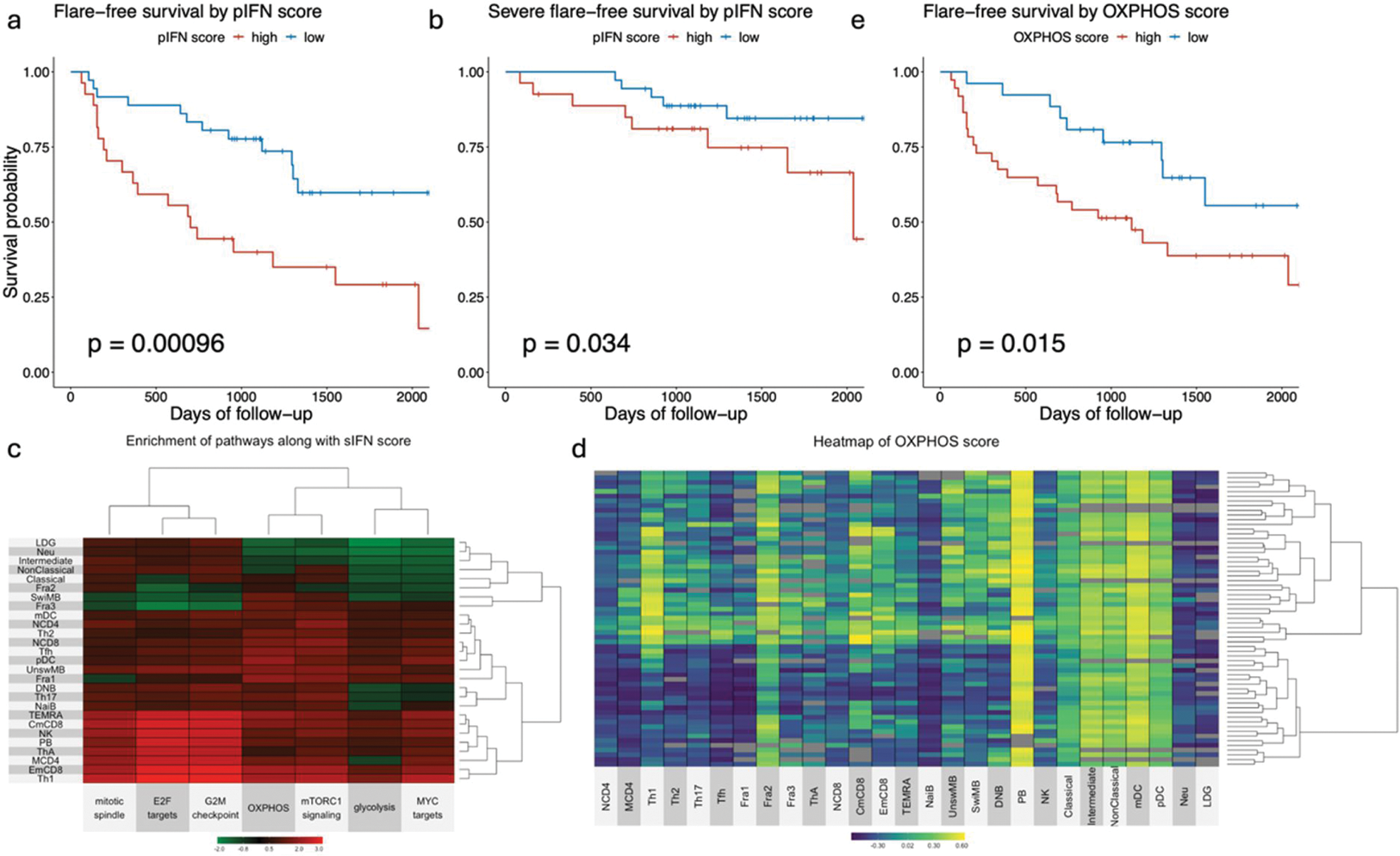
Results: A total of 63 and 18 patients were enrolled in the test and validation cohorts in this analysis, with a mean age of 46 ± 12 and 47 ± 21 years. 5 (7.9%) and 1 (5.6%) were male. The 3-year flare-free survival of each cohort was 61.6% ± 6.16% and 66.7% ± 11.1% with a median follow-up time of 1071 days and 1482 days, respectively. Weighted correlation network analysis revealed the interferon-stimulated genes (ISG) module was associated with subsequent flares across all immune subsets except neutrophil and low-density granulocyte (LDG). To calculate an interferon (IFN) score, we applied a scoring system previously reported based on RT-qPCR data [4] with some modifications to our bulk RNA-seq data. We defined a subset IFN score (sIFN score) as the IFN score of each subset of each patient and a patient IFN score (pIFN score) as the adjusted first component score using probabilistic principal component analysis for the sIFN scores. The pIFN score was highly correlated to the sIFN scores across all immune subsets (correlation coefficients ≥ 0.93). The subgroup with a high pIFN score showed poorer survival for flare and severe flare (Figure 1a and b). To uncover the pathways and the subsets associated with interferon signature, we performed a differential gene expression analysis with the sIFN score as a covariate. The gene set enrichment analysis showed that the pathways associated with cell cycle were enriched in Th1, memory CD8 subsets, and plasmablast along with the sIFN score (Figure 1c). Oxidative phosphorylation (OXPHOS) pathways tended to be enriched in T cells, B cells, and dendritic cells, but were suppressed in neutrophil, LDG, intermediate and non-classical monocytes as the sIFN score increased. We stratified the patients into 2 subgroups by hierarchical clustering based on the gene set variation analysis (GSVA) scores of OXPHOS. The high OXPHOS subgroup reflected high OXPHOS scores in Th1 and memory CD8 subsets, exhibiting poorer survival for flare (Figure 1d–e). Similar results were observed for cell cycle. By Lasso-Cox regression model analysis incorporating the pathway scores in Th1 and clinical variables like drug dosing, it was indicated that the OXPHOS score in Th1 had the most significant impact on survival for flare. In the validation cohort, these results were replicated.
Conclusion: The LLDAS patients with higher pIFN scores are more likely to experience subsequent flares, associated with the activation of OXPHOS and cell cycle in Th1 and memory CD8 subsets.
REFERENCES: [1] Ota M, et al. Cell. 2021;184(11):3006-3021.e17. doi:10.1016/j.cell.2021.03.056.
[2] Nakano M, et al. Cell. 2022;185(18):3375-3389.e21. doi:10.1016/j.cell.2022.07.021.
[3] Takeshima Y, et al. Ann Rheum Dis. 2022;81(6):845-853. doi:10.1136/annrheumdis-2021-221464.
[4] El-Sherbiny YM, et al. Sci Rep. 2018;8(1):5793. doi:10.1038/s41598-018-24198-1.
Flare-free survival by pIFN score and OXPHOS score
Acknowledgements: NIL.
Disclosure of Interests: Tatsuki Abe: None declared, Takahiro Itamiya TI has received grant/research support from Chugai Pharmaceutical., Yumi Tsuchida: None declared, Haruka Tsuchiya HT has received speaking fees and/or honoraria from Chugai Pharmaceutical., Hirofumi Shoda HS has received speaking fees from Chugai pharmaceutical., Tomohisa Okamura TO has received speaking fees from Bristol Myers Squibb., TO has received grant/research support from Chugai Pharmaceutical., Keishi Fujio KF has received speaking fees from Chugai Pharmaceutical., KF has received grant/research support from Chugai Pharmaceutical.