

Background: Pulmonary arterial hypertension (PAH) is a devastating complication of systemic sclerosis (SSc), as it represents one of the leading causes of mortality. The DETECT algorithm is a screening tool to identify patients with SSc-PAH in the asymptomatic stage. This algorithm shows an optimal performance identifying almost all cases of PAH in SSc patients (over 90%) but suffers from a low specificity (less than 50%). Consequently, in a significant proportion of patients, the diagnosis of PAH is not confirmed after right heart catheterization (RHC), a costly and invasive procedure necessary for diagnosing PAH.
Objectives: We aimed to discover new blood-based biomarkers that consistently recognize SSc patients at risk of PAH, to improve current screening tools and to reduce the number of unnecessary RHC evaluations.
Methods: To deeply screen changes in secreted protein abundances, we performed a label-free quantitative proteomic analysis by liquid chromatography with tandem mass spectrometry (LC-MS-MS) on 47 serum samples from patients with SSc. All patients were considered at risk of PAH according to the DETECT algorithm. After RHC evaluation, PAH was diagnosed in 16 subjects (SSc-PAH group), while the other 31 subjects showed normal haemodynamic values (SSc-nonPAH group) according to the 2022 ESC/ERS definition. We used linear regression to identify which differentially abundant proteins associated with higher pulmonary vascular resistance (PVR) or mean pulmonary arterial pressure (mPAP), corrected for sex, age, BMI, use of statins and immunosuppressants. Logistic regression was used to test the performance of a panel of six selected proteins in classifying the two groups of patients. Public single-cell RNA sequencing datasets (scRNAseq) were used to investigate the expression of the six selected proteins in lung samples from patients with SSc-PAH and healthy controls 1 .
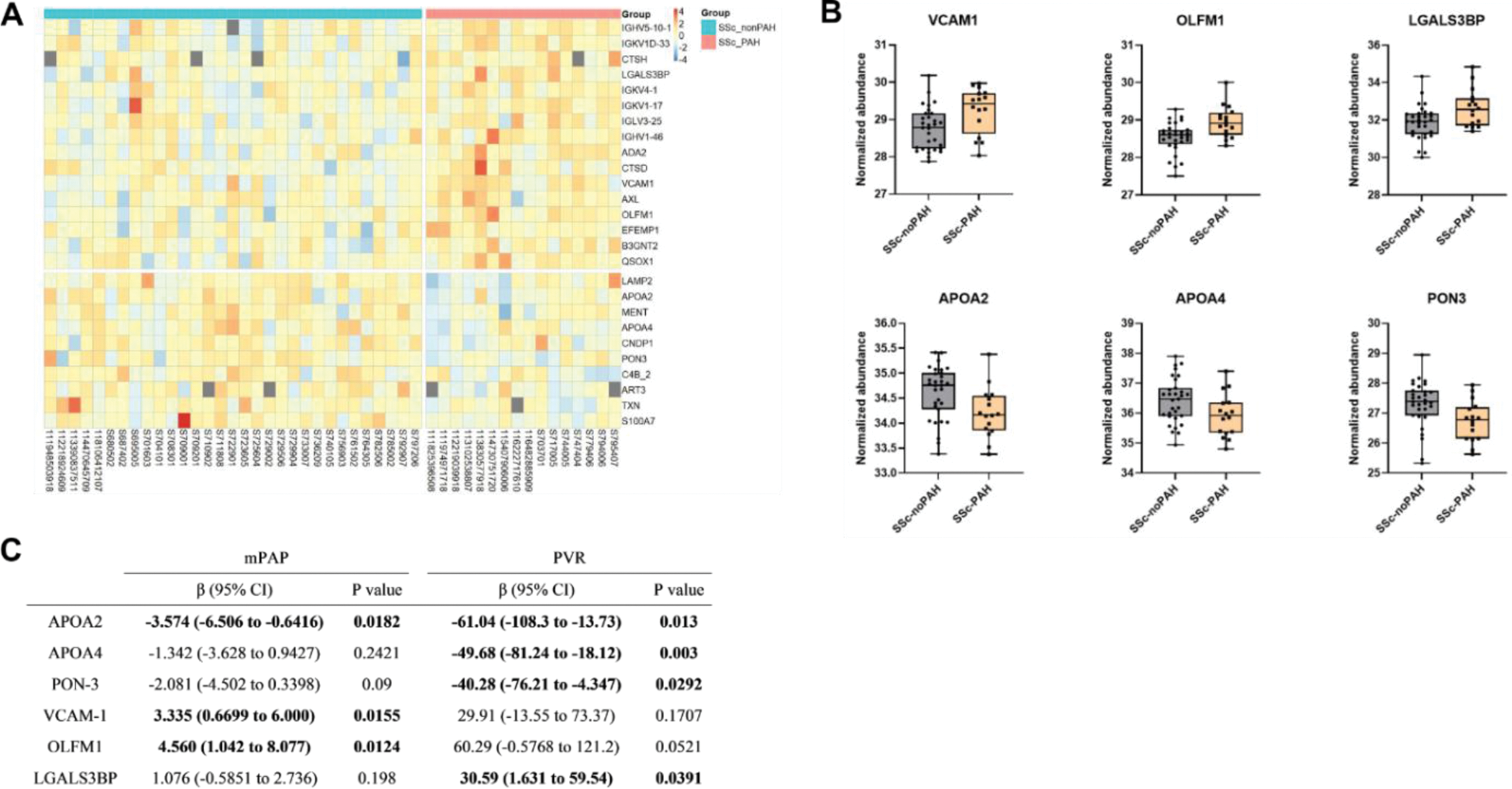
Results: The proteomic analysis provided 26 proteins differentially abundant between the two groups (p-value < 0.05) (Figure 1A-B). Sixteen of them were increased in the SSc-PAH group. Galectin-3 binding protein (LGALS3BP), paraoxonase-3 (PON-3), apolipoproteins A2 and A4 (APOA2 and APOA4) showed a significant association with PVR. APOA2, olfactomedin-1 (OLFM1) and vascular cell adhesion protein 1 (VCAM-1) showed a significant association with mPAP (Figure 1C). We focused the subsequent analysis on these six proteins. Using logistic regression, the model including the six proteins was able to discriminate SSc-PAH from SSc-nonPAH patients with an area under the receiver operating characteristic curve (ROC-AUC) of 0.857 (95% CI: 0.753-0.960). For technical validation of these results, serum levels of the candidate biomarkers were analysed by ELISA. Additionally, we analysed public scRNAseq datasets of lung tissue samples from SSc-PAH patients and healthy controls. VCAM-1 and OLFM-1 had significantly higher expression in SSc patients compared to controls in endothelial cells (p < 0.001 and FC = 1.65 for VCAM1, p < 0.001 and FC = 1.44 for OLFM-1).
Conclusion: In this preliminary analysis, we identified six candidate biomarkers that could contribute to identification of SSc patients at risk of PAH. Dysfunctional lipoprotein metabolism has been shown to be involved in PAH, supporting a potential role of APOA2, APOA4 and PON3. The increased expression of VCAM-1 and OLFM1 in endothelial cells of SSc patients also supports their relevance in the context of SSc-PAH. These preliminary findings will be validated in external cohorts and the biological relevance of the candidate biomarkers will be further investigated, focusing in particular on the added value to the DETECT algorithm, the currently recommended screening tool.
REFERENCES: [1] Papazoglou, A. et al. Arthritis Rheumatol. 2022.
A) Heatmap of the normalized abundance of the 26 differentially abundant proteins. Proteins are ordered according to difference in normalized abundance. B) Normalized abundance of the 6 selected proteins. Comparisons are performed with an unpaired t-test. C) Multiple linear regression analysis between candidate biomarkers and haemodynamic parameters.
Acknowledgements: We thank the Functional Genomics Center Zurich for the excellent support for the proteomic analysis.
Disclosure of Interests: Gino Andrea Bonazza: None declared, Cosimo Bruni: None declared, Kristina Buerki: None declared, Silvia Ulrich: None declared, Mona Lichtblau: None declared, Oliver Distler Speaker fee: Bayer, Boehringer Ingelheim, Janssen, Medscape, Consultancy fee: 4P-Pharma, Abbvie, Acceleron, Alcimed, Altavant Siences, Amgen, AnaMar, Arxx, AstraZeneca, Baecon, Blade, Bayer, Boehringer Ingelheim, Corbus, CSL Behring, Galapagos, Glenmark, Horizon, Inventiva, Kymera, Lupin, Miltenyi Biotec, Mitsubishi Tanabe, MSD, Novartis, Prometheus, Redxpharna, Roivant, Sanofi and Topadur, Research Grants: Kymera, Mitsubishi Tanabe, Boehringer Ingelheim, Przemyslaw Blyszczuk: None declared, Gabriela Kania: None declared.