

Background: Rheumatoid Arthritis (RA) is very heterogeneous, yet disease discerning patterns remain largely elusive, thereby impeding the progress of aetiological research and the development of novel treatment strategies. The recent surge in data-driven techniques[1] however, harbors great potential for stratifying complex diseases by unlocking the potential of big data sources such as the Electronic Health Records (EHR).
Objectives: Our aim was to identify clinically relevant phenotypic subgroups from a DMARD-naive RA population based on clinical features available at baseline.
Methods: 1,387 RA patients were collected from the rheumatology outpatient clinic[2], for which we constructed patient specific profiles from lab information (serology (ACPA/RF), hematology workup), demographics (age, sex) and joint inflammation patterns at baseline. We employed a deep learning strategy to cluster the patients into phenotypically distinct patient subsets. Using an independent validation set (n=769) we extensively evaluated the robustness and transferability of the proposed patient stratification. Moreover, associations with methotrexate (MTX) response, remission (DAS44<1.6) after 1 year, and known RA genetic variants were evaluated, to assess the potential clinical and aetiological relevance of the identified patient subsets.
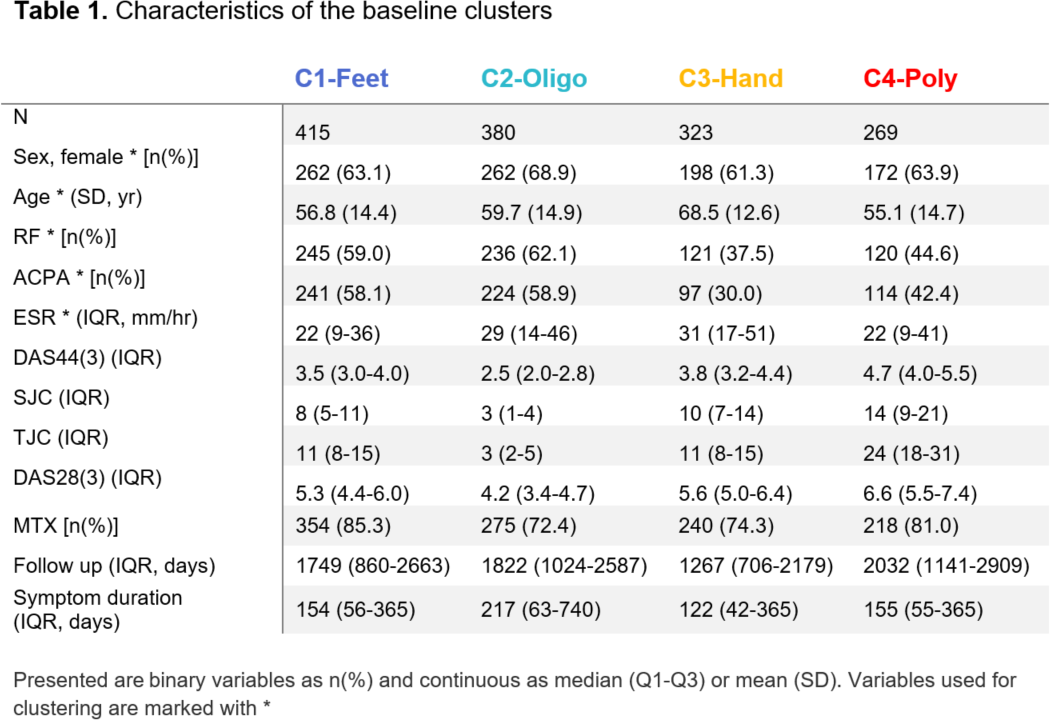
Results: We detected four baseline clusters (Table 1; Figure 1A):
Cluster 1 - seropositive arthritis in feet,
Cluster 2 - seropositive oligoarticular disease,
Cluster 3 - seronegative elderly patients with arthritis in the hands,
Cluster 4 - seronegative polyarthritis.
Robustness analyses revealed that the clusters were stable (80% of patients recurrently co-clustered across 1000 random permutations of the data), not biased by physicians and replicated in separate data.
The MTX failure rate (Figure 1B) significantly differed across clusters (I to IV): 27%, 23%, 16%, 30%, respectively (P<0.001). The hands subset (C3) was significantly more likely to stay on MTX than polyarthritis - (C4; 0.48 (95%CI 0.35-0.77), P<0.001) as well as the feet subgroup (C1; HR:0.55 (0.37-0.82), P=0.003). Interestingly, the interaction between ACPA and treatment failure differed between clusters (Figure 1C). Furthermore, the remission rates (Figure 1D) for the four clusters were 44.3%, 47.4%, 55.7%, 38.5% (P=0.007), showing a similar pattern as the MTX analysis, with C3 being more likely to reach remission than C4 (HR 1.65 (95%CI 1.2-2.29), P=0.002). When testing clusters for enrichment of known RA genetic (risk) variants, we found 9 single nucleotide polymorphisms (SNPs) to be enriched for the different clusters: C1) IL2RA, SPRED2, RUNX1, C2) COG6, GRHL2, ETS1, C3) AFF3 and C4) CCL21, CTLA4.
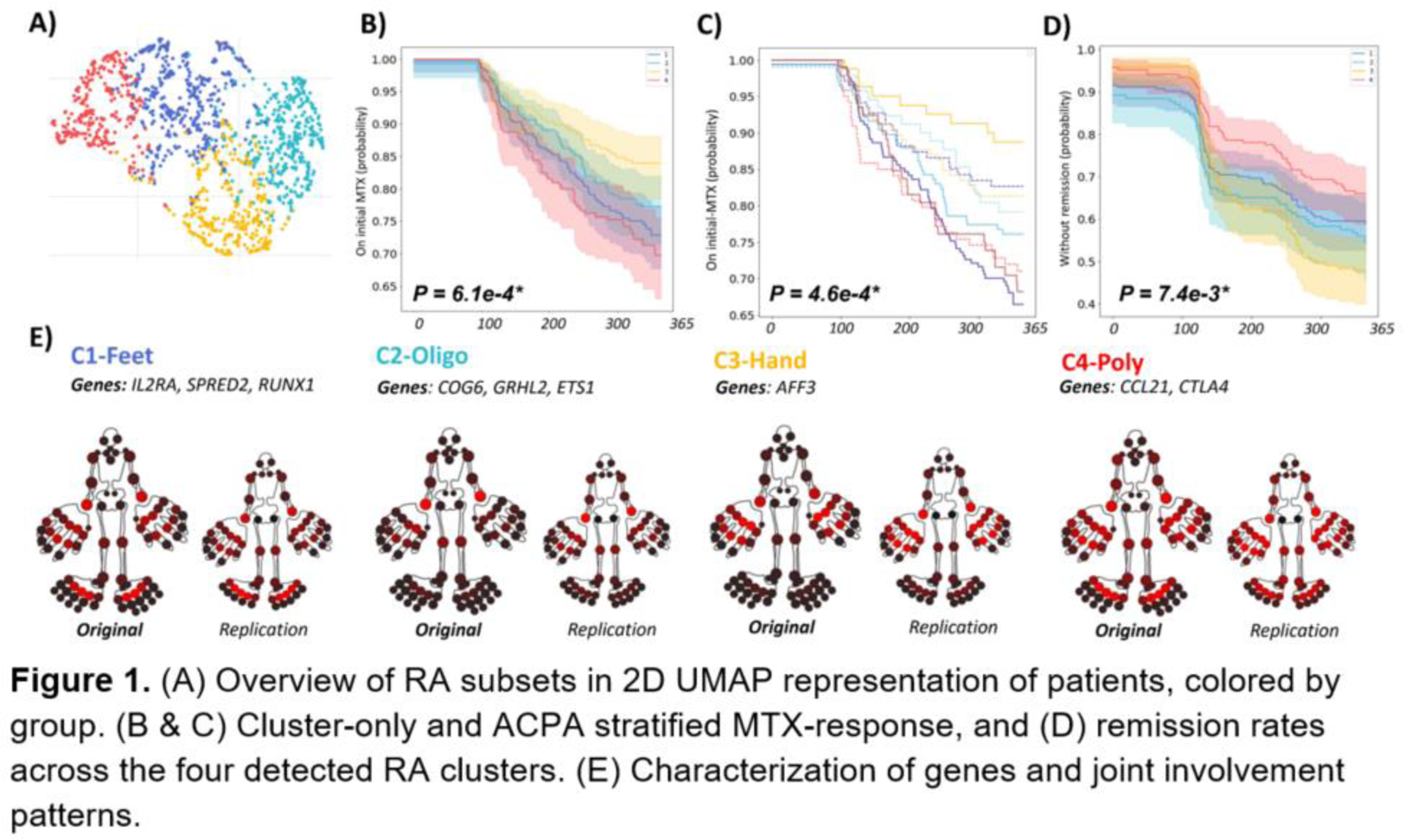
Conclusion: Based on routinely collected diagnostic baseline data, we identified four clusters that differed in clinical outcomes. Of particular note are the hands- and feet clusters, and the interactions with SNPs that suggested different genetic risk profiles. Our identification of replicable and clinically relevant subsets of RA demonstrates the value of data-driven techniques to identify hidden structure.
REFERENCES: [1] Maarseveen TD et al. doi: 10.2196/23930
[2] Ahlqvist E, et al. doi: 10.1016/S2213-8587(18)30051-2
[3] Böhringer, S. and Lohmann, D. doi:10.1016/j.jspi.2021.07.013
Acknowledgements: This project has received funding from the EIT health body for DigiPrevent (funded in April 2022) and from the Horizon Europe program under grant agreement no. 101095052 (SQUEEZE) and no. 10108071 (SPIDERR). The study has further received funding by the ZonMW Klinische Fellow No. 40-00703-97-19069, as well as the ZonMW Open Competitie, No. 09120012110075.
Disclosure of Interests: None declared.