

Background: High-throughput protein microarrays are increasingly employed to investigate disease-related autoantibodies in patients with SLE for identification of novel serum autoantibodies and their signatures that correlate with and potentially predict disease activity, flare and damage in SLE. Herein, we present extended analyses of our original investigation [1] into novel autoantibodies in predicting disease activity and flares in patients with SLE.
Objectives: To validate our preliminarily discovered putative autoantibodies in a larger cohort of SLE patients and correlate these autoantibodies with clinical manifestations, disease activity and damage longitudinally.
Methods: Serum autoantibody profiling of 60 SLE patients was compared with 15 healthy subjects using the Sengenics I-Ome high-throughput proteomic microarray that can identify over 1,600 proteins in the sera. We conducted linear modeling analyses to identify novel autoantibodies that were significantly altered in SLE patients compared with healthy subjects. We also determined whether these autoantibodies could predict the baseline risk of disease flares, organ involvement, and SLEDAI-2K at 3 and 6 months from baseline.
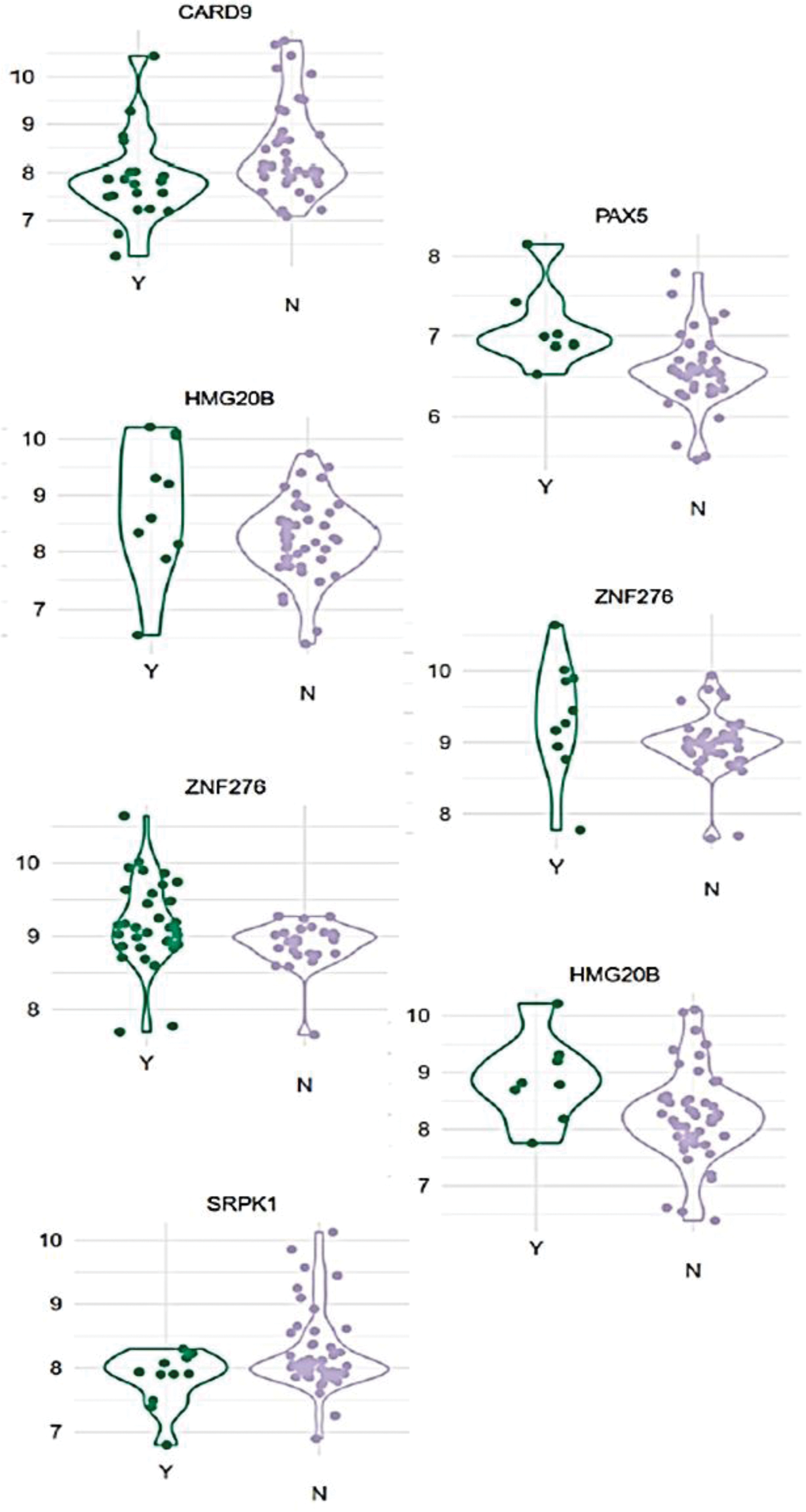
Results: Bioinformatics analyses identified several autoantibodies weighted by their occurrence frequencies in flares, organ involvement, and correlation with disease activity status (Figure 1). Compared to healthy subjects, autoantibodies against CARD9 (p=0.019), PAX5 (p= 0.018), HMG20B (p=0.05), and ZNF276(p=0.019) were associated with baseline and subsequent risk of overall disease flares, renal involvement, and vasculitis. CARD9 is an adapter of C-type lectin receptors with a caspase-recruitment domain that recruits BCL10 (a regulator of apoptosis and NFkB/MAPK-mediated signaling). PAX5 is a transcription factor that regulates B-cell responses. HMG20B facilitates DNA binding,and positively regulates neuronal differentiation. ZNF276 facilitates dsDNA binding and regulates transcription. Anti-SRPK1,regulates splicing, was predictive of CNS involvement. The serum levels of HNRNPUL1(p=0.049), ZNF323(p=0.011), and HMG20B were significantly associated with higher SLEDAI-2K at 3 and 6 months. Additionally, HNRNPA2/B1, a nuclear DNA sensor that amplifies interferon-1 responses, correlated with higher SLEDAI-2K at 6 months (p= 0.0035). The five most common autoantibody signatures in extended analysis targeted SSB, CCNB1,TROVE2, DEK, and HNRNPA2B1, which were increased in SLE patients compared to healthy subjects.
Violin plots of representative antigens with p-value < 0.05 from Linear Modelling Analysis
Abbreviations:
CARD9: Caspase recruitment domain-containing protein 9 is an adaptor protein encoded by the CARD9 gene, mediates signals from pattern recognition receptors, and regulates inflammation.
PAX5 : Paired box protein -5 is a key regulator in B cell development at different stages, involved in B-cell signaling and adhesion and migration of B lymphocytes.
HMG20B : High Mobility Group 20B is a Protein Coding gene that enables DNA binding and regulates gene expression and neuronal differentiation.
ZNF276: Zinc Finger Protein 276 is a Protein Coding gene that enables sequence-specific double-stranded DNA binding activity.
SRPK1: Serine/Arginine-Rich Protein-Specific Kinase 1 protein regulates the expression of apoptosis-related genes.
Y denotes presence of autoantigen in serum samples.
N denotes absence of autoantigens in serum samples.
Conclusion: Using high-throughput proteomic microarray, a number of novel autoantibodies and their signatures related to disease activity and specific organ involvement were identified in SLE patients. Further validations of the pathophysiology of these novel autoantibodies detected in SLE patients with larger cohorts internationally are required.
REFERENCES: [1] Mak A, Kow NY, Ismail NH, Anuar ND, Rutt NH, Cho J, et al. Detection of putative autoantibodies in systemic lupus erythematous using a novel native-conformation protein microarray platform. Lupus. 2020;29(14):1948-54.
Acknowledgements: NIL.
Disclosure of Interests: None declared.