

Background: Sjögren’s disease (SjD) is a systemic autoimmune disorder that primarily affects the exocrine glands, particularly the salivary and lacrimal glands. Recent efforts have exploited serum and saliva metabolome analysis to enhance our understanding of pathogenetic mechanisms and the development of novel biomarkers, however these studies are hampered by limited patient numbers, poorly characterized SjD cases, the absence of proper control groups, a lack of clinicohistologic associations, and variations in the metabolomic analysis techniques used.
Objectives: The aim of the current study is to enlighten the latent biochemical background of SjD and to identify potential biomarkers facilitating early non-interventional SjD diagnosis.
Methods: To investigate metabolomic profiles associated with SjD, we conducted a comprehensive analysis using saliva samples from patients presenting with sicca symptoms. All patients diagnosed with SjD fulfil the 2016 ACR-EULAR classification criteria and all minor salivary gland biopsies were re-evaluated by the same expert on SjD salivary gland pathology. Patients not fulfilling the criteria for SjD were classified as sicca controls (n=21 for QTOF and 17 for 1H NMR). Our metabolomics profiling protocol allows for a simultaneous, non-targeted measurement of a wide variety of small molecules, exploiting 2 different techniques: high-resolution mass spectrometry (QTOF) and high-resolution 1H 1D NMR spectroscopy. Patients were categorized based on their focus score (FS) into low (LFS, FS<1, n= 8 f for QTOF and 6 for 1H NMR), medium (MFS, 1<FS<3, n=18 for QTOF and 15 for 1H NMR), and high focus score (HFS, FS>3, n= 9 for QTOF and 9 for 1H NMR) groups. In our analysis, we considered potential influences of gender, age, saliva flow rate during sample collection, and FS. Multi- and univariate statistical methodologies were applied for group classification and biomarker detection. Partial Least Squares Projection to Latent Structures modeling (PLS; SIMCA v. 14, MKS Umetrics AB) was used to correlate spectroscopic data with Focus score values. MetaboAnalyst, a web-based platform for metabolomic data analysis and interpretation was also used.
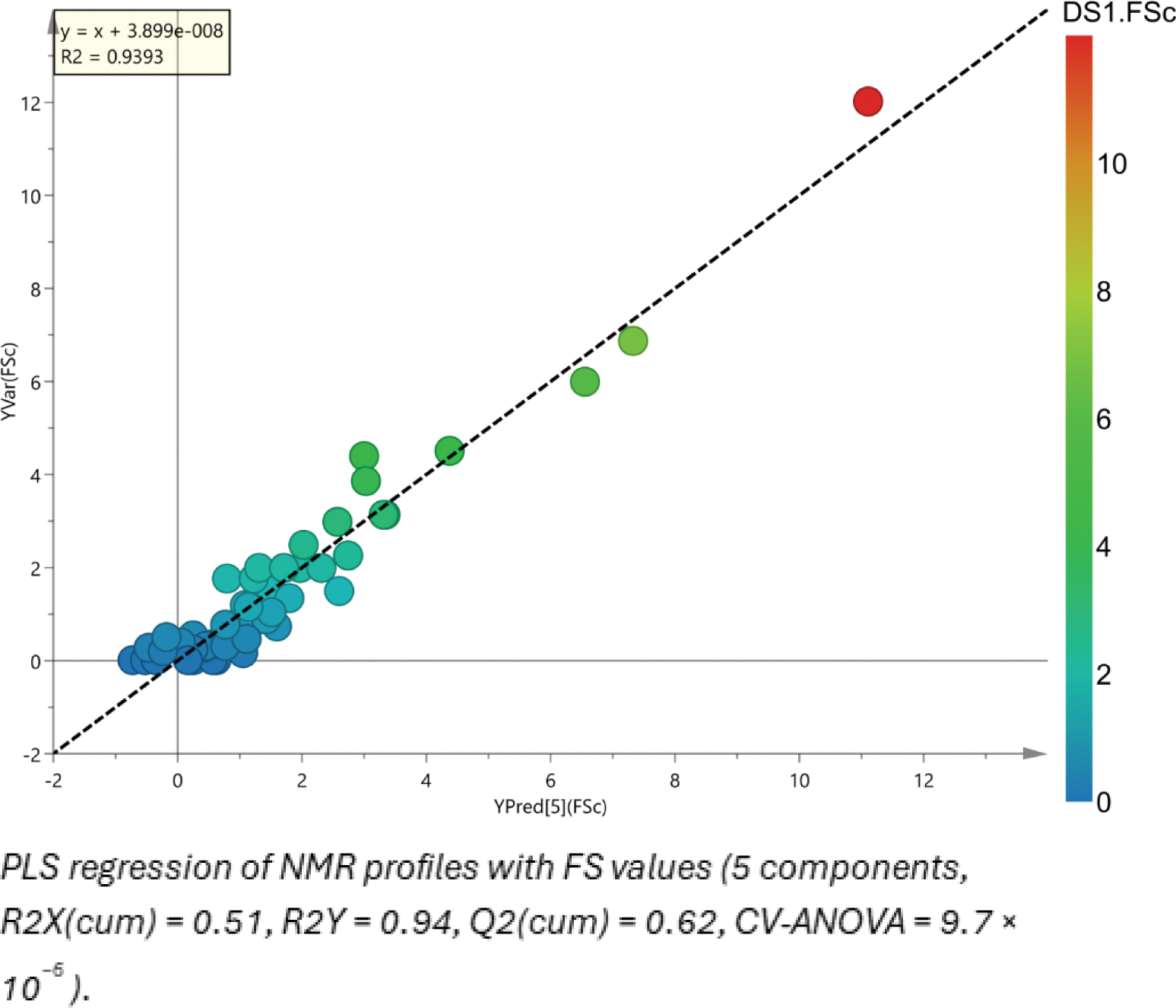
Results: Utilizing multivariate statistical methodologies, patients were successfully categorized into Low, Moderate, and High Focus Score (LFS, MFS, HFS) groups, as well as into Sjögren and Non-Sjögren diagnostic categories, demonstrating acceptable model performance. Preliminary analyses identified upregulated metabolites in Sjögren patients saliva, including Choline (Fold Change, FC=1.3), Lactic acid (FC=1.4), Taurine (FC=1.1) from the 1H NMR analysis, and Neuraminic acid (FC=2.2), Deoxyinosine (FC=2.8), Pimelylcarnitine (FC=1.8) from the QTOF analysis. In HFS patients, elevated levels of Leucine (FC=1.3), Indoleacrylic acid (FC=1.2), Guanidinoacetic acid (FC=1.3) were observed, while Diaminopropionic acid (FC=1.4) and Prostaglandin E3 (FC=1.3) were decreased. Proof of concept correlation between the metabolic fingerprint and focus score values revealed a high correlation (R2=0.94) with strong statistical significance (p=9.7 × 10−6).
Conclusion: To the best of our knowledge, this is the first study investigating saliva sample metabolomics with 2 different techniques, coupled with biopsy FS data. This approach provides an insight into the distinctive alterations characterizing both sicca controls and Sjögren’s disease patients across different disease stages. According to the findings the intensity of inflammation, as depicted by the salivary biopsy focus score, is highly associated with the salivary metabolome landscape. Meanwhile some metabolites presen in the saliva have emerged as novel biomarkers aiming to offer a non-invasive tool to support SjD clinical diagnosis and histologic classification. Further studies are needed to validate these results and establish the clinical utility in SjD.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.