

Background: Stratification of patients affected with systemic autoimmune diseases according to clinical and endotypic features may provide valuable insights to disease mechanisms and expected response to target treatments. Belimumab is increasingly used as add-on therapy in refractory systemic lupus erythematosus (SLE), yet the heterogeneity of patients undergoing belimumab was not thoroughly investigated. Unsupervised machine learning allows cluster profiling through unbiased assessment of multiple variables in the model.
Objectives: In this study we aimed at exploring clusters defined by clinical and/or laboratory features across patients initiating belimumab due to refractory SLE, and to investigate possible relationships with clinical outcome at different timepoints.
Methods: We analyzed data from the Italian multicentric SLE database (LIRE – Lupus Italian REgistry) encompassing prospectively collected records of patients commencing a novel treatment due to refractory SLE. We selected patients initiating belimumab. Multiple Factor Analysis for Mixed Data (FAMD) was employed to differentiate patient clusters. To define the number of clusters, Hierarchical Clustering on Principal Components was performed using Ward’s criterion and a Euclidian distance metric on the selected FAMD principal components. Inter-cluster inertia gain was used to determine the optimal division level.
Disease activity was measured by SLE-disease activity index (SLEDAI-2K), damage by the SLICC damage index (SDI). Clinical response was defined as clinical (c)SLEDAI-2K=0 and was evaluated at 6 and 12 months (T6 and T12). Logistic regression analysis was performed to assess clustering effect on response rates. Significance was set at 5%.
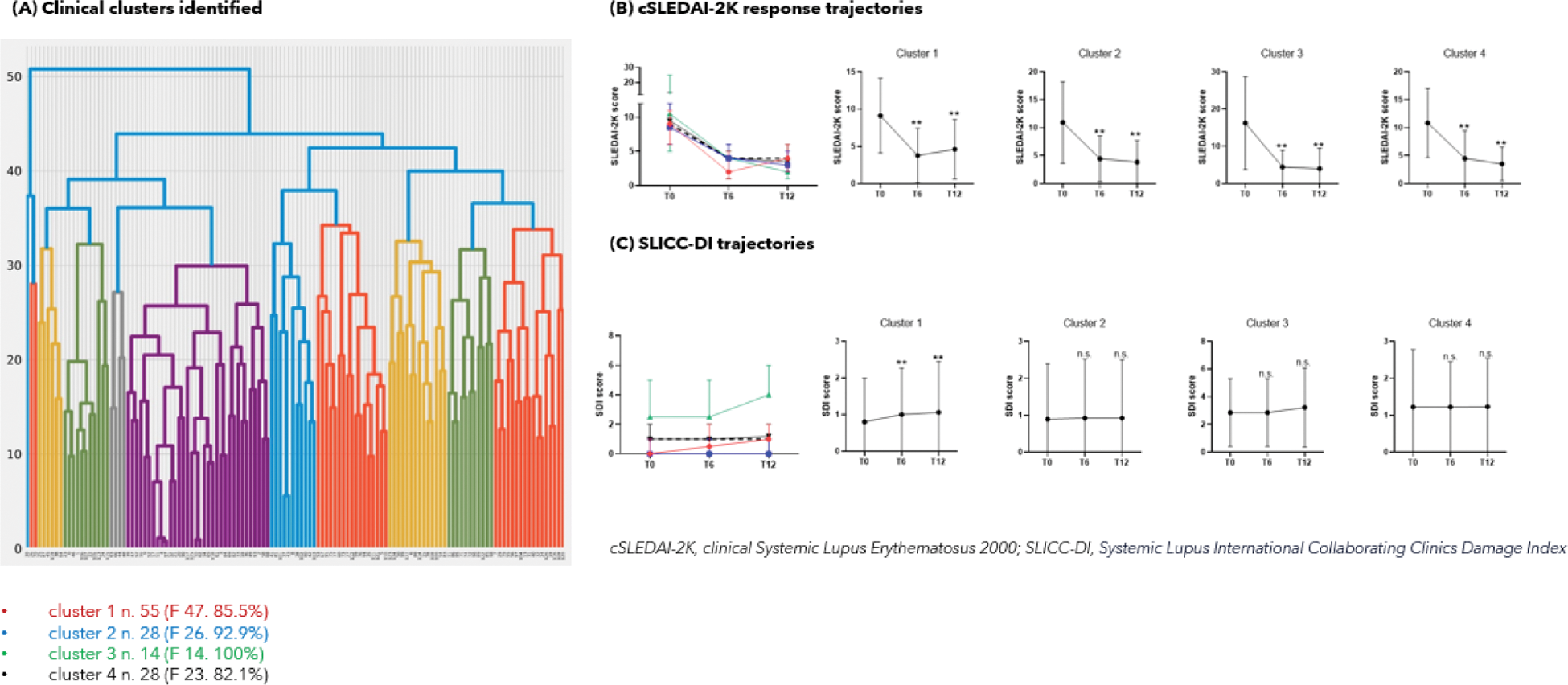
Results: Hierarchical clustering identified five principal components recapitulating 4 main clusters from 125 SLE patients initiating belimumab ( Figure 1A ). Cluster 2 (n=28) included patients with renal involvement and enhanced complement consumption while Cluster 3 (n=14) exhibited activity in neuropsychiatric and cardiovascular domains. Cluster 3 displayed the highest baseline SDI (p=0.006) enriched in neurological and cardiovascular items (p<0.001), and the lowest baseline hydroxychloroquine intake (78.6% vs. 95.2%, p=0.05). Cluster 4 (n=28) gathered patients with haematological abnormalities, while Cluster 1 (n=55) included patients showing no identifiable disease pattern yet entailing a lower SDI.
Response trajectories throughout T6 and T12 were comparable across clusters, with Cluster 2 showing a numerically higher proportion of cSLEDAI-2K responders at T12 (70.8% vs. 52.3%, p=0.082) ( Figure 1B ). Clustering did not predict clinical response. SDI at T6 and T12 was significantly greater in Cluster 3 (p<0.01 for both), yet damage accrued significantly within Cluster 1 throughout month 12 (0 (0-2) vs. 1 (1-2), p<0.001) ( Figure 1C ).
Conclusion: Unsupervised clustering highlighted the clinical heterogeneity of patients candidate to belimumab due to refractory SLE. Although clustering did not clearly predict clinical outcome in this cohort, a greater response rate seems likely in patients with active serology and renal involvement. Combining expert judgement with machine learning may enable optimized eligibility and prediction of response upon belimumab treatment.
REFERENCES: NIL.
Acknowledgements: Prof. Antonella Zambon, Anna Zanetti, Isabella Sala (University Milan Bicocca); Greta Carrara (Centro Studi SIR).
Disclosure of Interests: Mariele Gatto GSK, AZ, Janssen, Vincenzo Venerito: None declared, Davide Rozza: None declared, Gianpiero Landolfi: None declared, Carlo Alberto Scirè: None declared, Gian Domenico Sebastiani: None declared, Fabrizio Conti: None declared, Andrea Doria GSK, AZ, Celgene, Eli Lilly, Roche, Pfizer, BMS, GSK.