

Background: Around 3% of the global population suffers from Fibromyalgia (FM) and the average diagnostic delay is six years [1]. It is a hard disease to diagnose as it has symptoms that are common in non-Inflammatory and inflammatory diseases [2]. An outcome classification based on self-reported symptoms could benefit patients, by allowing patients to be referred to specialists sooner. “Rheumatic?” is an online symptom checker originally designed to differentiate patients with immune-mediated from non-immune-mediated rheumatic disease [3], yet it might also aid in the early identification of patients at risk of FM. A previous machine-learning approach trained on “Rheumatic?” data performed moderately well at diagnosis prediction task, with an AUROCS of 0.54 – 0.79 [3].
Objectives: We investigate whether self-reported symptoms collected from “ Rheumatic?” can be used to identify FM diagnosis.
Methods: “Rheumatic?” includes a baseline questionnaire about current symptoms and past diagnoses, along with three follow-up surveys (at 3,6 and 12 months) about healthcare visits, medication, and further diagnoses. We selected participants that entered the “Rheumatic?” between July 2021 and July 2022.
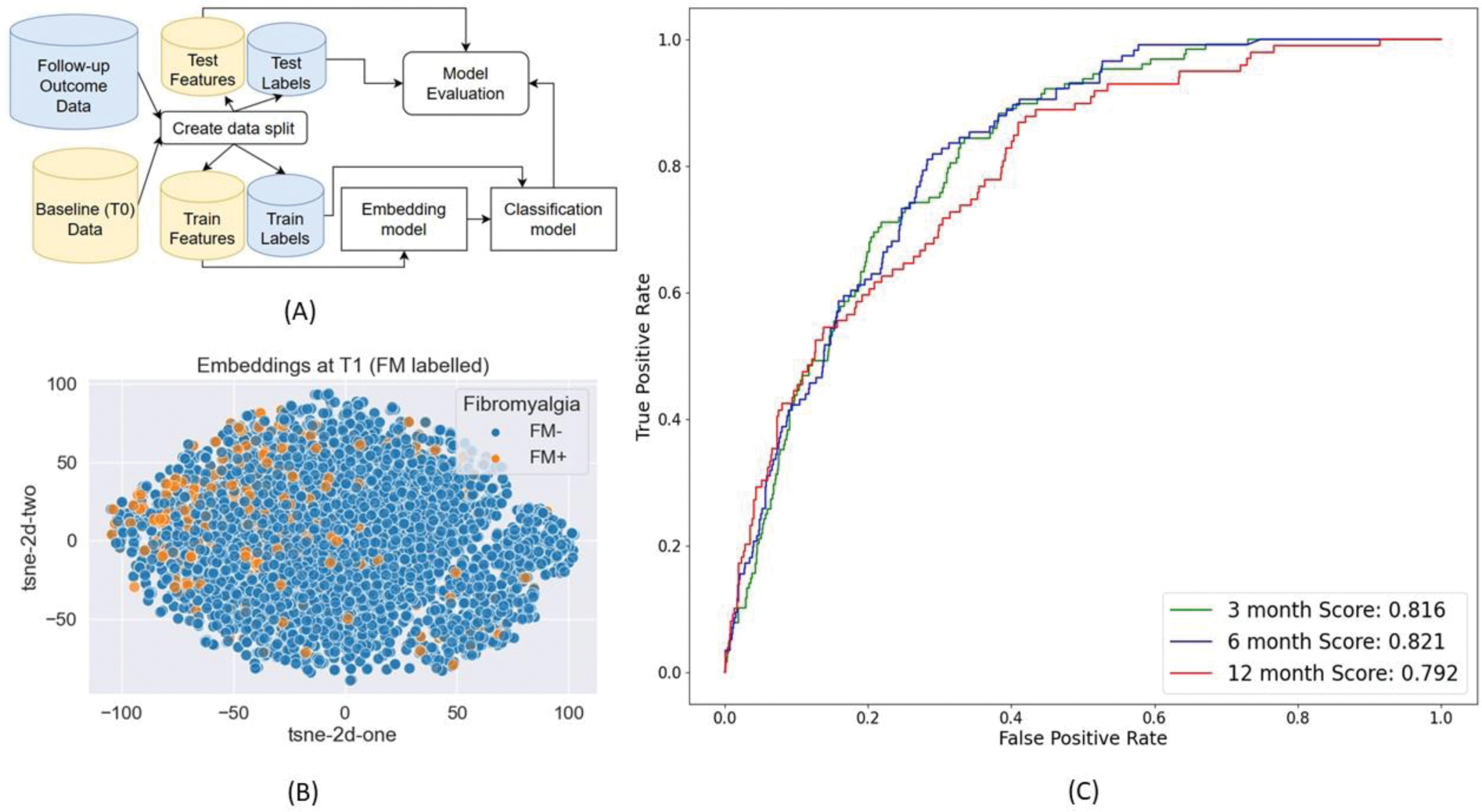
Each participant’s baseline data was projected into an embedding (Figure 1A) using a pre-trained auto-encoder model. The embedding was then used, along with FM outcomes as labels, to train an XG-Boost classifier (Figure 1B).
The data was split 80:20 into a train set and test set. The train set was used to train the auto-encoder model in an unsupervised manner, and the classifier in a supervised manner. The test set was used to get the final model performance metrics. We used the Area Under the Receiver Operating Curve Score (AUROCS) to assess model performance. A separate pipeline was created for every follow-up timepoint.
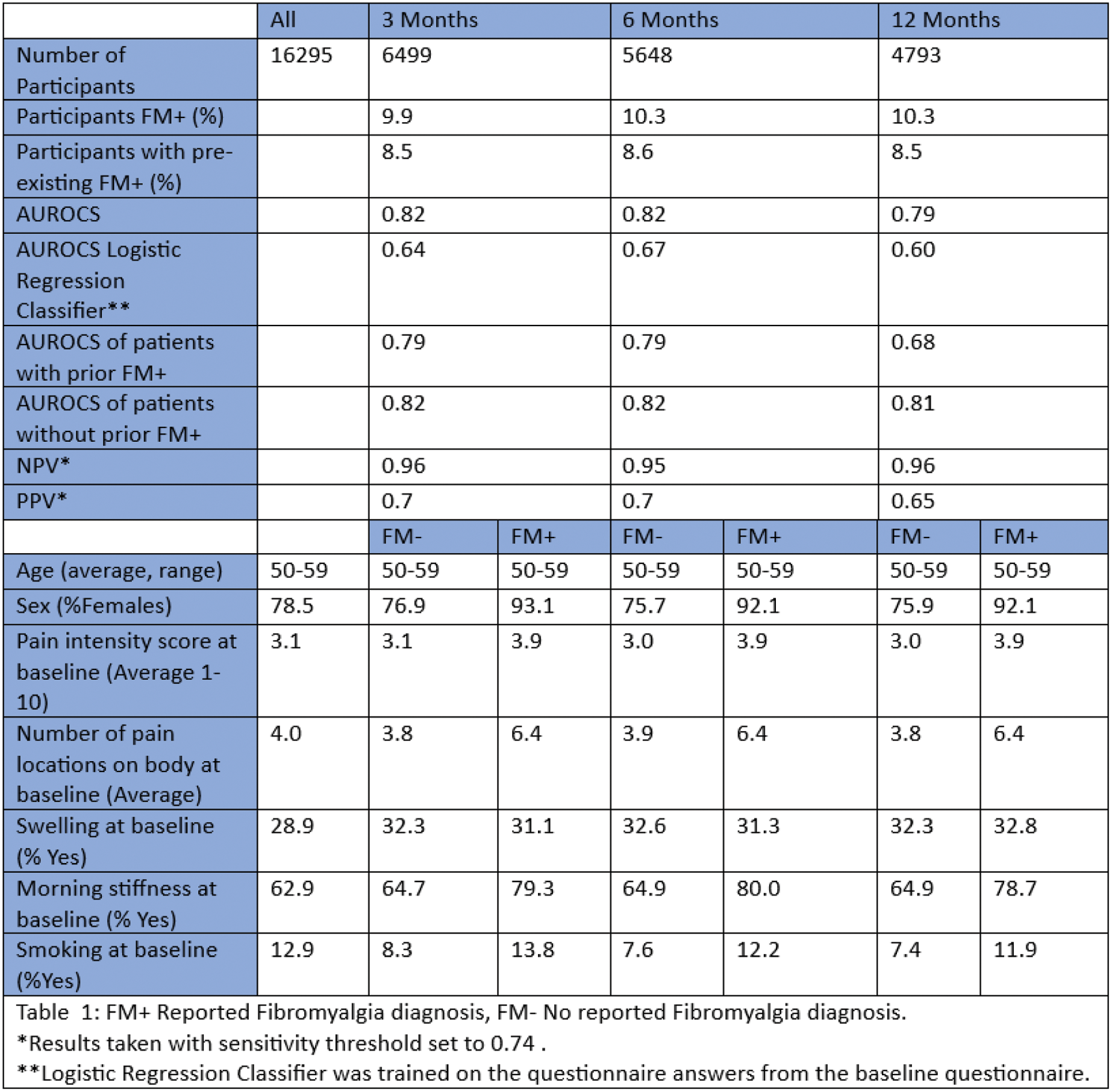
Results: The number of participants at each follow-up changed as not every participant replied to a follow-up survey (Table 1). As expected, participants who reported a FM diagnosis were majorly women, reported more intense pain and reported pain in more areas of the body compared to those without FM. In contrast to the known (textbook) disease pattern, FM patients more frequently reported morning stiffness and swelling.
Our algorithm performs well for predicting FM diagnosis at all follow-up timepoints with overall AUROCS never falling below 0.79 (Figure 1C). Performances decline with increasing time between baseline and follow-up. This is to be expected as the number of participants available for training decreases, and the time between reported symptoms and diagnosis increases. Model performance was highly comparable between patient groups with and without pre-existing FM diagnosis, with performances being slightly better for patients without pre-existing FM diagnosis.
Conclusion: We develop an algorithm to embed symptom checker questionnaire data and perform self-reported FM diagnosis classification using that embedding. Our pipeline obtained a consistently high performance. Such a pipeline can be useful for clinicians by allowing faster diagnosis based on symptom checker responses from patients.
REFERENCES: [1] Queiroz. Pain Headache Rep (2013). 10.1007/s11916-013-0356-5
[2] Gendelman et al. Best Pract Res Clin Rheumatology (2018). 10.1016/j.berh.2019.01.019
[3] Knevel et al. Front Med (2022). 10.3389/fmed.2022.774945
. A) Overview of Fibromyalgia diagnosis algorithim. B) 2d representation of the baseline questionnaire data embedding. The embedding was generated using the latent space of the trained auto-encoder. FM+: Participants with self-reported Fibromyalgia diagnosis. FM-: Participants who did not report this diagnosis. C) Reciever Operating Characteristc Curve, along with the Area Under Curve Values of Fibromyalgia diagnosis prediction performance
Acknowledgements: This project has received funding from Horizon Europe programme under grant agreement no. 10108071 (SPIDERR).
Disclosure of Interests: Daniyal Selani: None declared, Ling Qin: None declared, Floor Zegers: None declared, Lars Klareskog: None declared, Ewout Steyerberg: None declared, Saskia Le Cessie: None declared, Barbara Axnäs Elsa Science is SaMD Company, Marcel J.T. Reinders: None declared, Erik B. van den Akker: None declared, Rachel Knevel: None declared