

Background: Lupus nephritis (LN) is a common complication of Systemic Lupus Erythematosus often carrying high mortality and morbidity. Fibrosis in LN has been detected in approximately 40% of adults with SLE [1], often accompanied by tubular atrophy. Even though glomerular injury has been the focus in the pathogenesis of LN, interstitial fibrosis is thought to be more indicative of severity injury and progression to renal failure [2], warranting further research on targets and mechanisms of interstitial fibrosis in LN. In this study, we analyzed co-expressed pathways in the pathogenesis of renal fibrosis in the context of LN using bioinformatics software.
Objectives: We aimed to identify co-expressed genes starting from overrepresented genes in the human phenotype of renal fibrosis in samples of kidney biopsies.
Methods: We downloaded the dataset GSE112943 from the GEO platform. This dataset contains 14 samples of kidney biopsies from patients with LN and 6 samples of healthy controls. We utilized the Gene Set Enrichment Analysis (GSEA) tool to identify overrepresented genes in the phenotype of renal fibrosis, which was downloaded from the Human Collection of the Molecular Signatures Database. To identify co-expressed genes, we input the overrepresented genes in the Genemania platform. We performed an ontological analysis on the co-expressed genes with the Database for Annotation, Visualization and Integrated Discovery (DAVID). We assessed protein-protein interactions (PPIs) using the StringApp via Cytoscape.
Results: 6 genes were enriched in the cohort of patients with LN, while 20 genes were found to be co-expressed. Ontological analysis found that these genes participate in pathways related to apoptosis (Table 1). PPIs are shown in Figure 1.
An ontological analysis performed in the DAVID platform, showing the biological processes involving the co-expressed genes.
| Ontological Analyisis: Biological Processes | ||
|---|---|---|
| Category | Genes | P-Value |
| Maintenance of protein location in nucleus | ARL2BP, ARL2 | 1,0E0 |
| Positive regulation of release of cytochrome c from mitochondria | TNFSF10, PDCD5 | 1,0E0 |
| Positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | TNFSF10, PDCD5 | 1,0E0 |
(A) Shows overrepresented genes in the phenotype of renal fibrosis in the inner circle, and co-expressed genes in the outer circle. (B) Shows PPIs among the co-expressed genes.
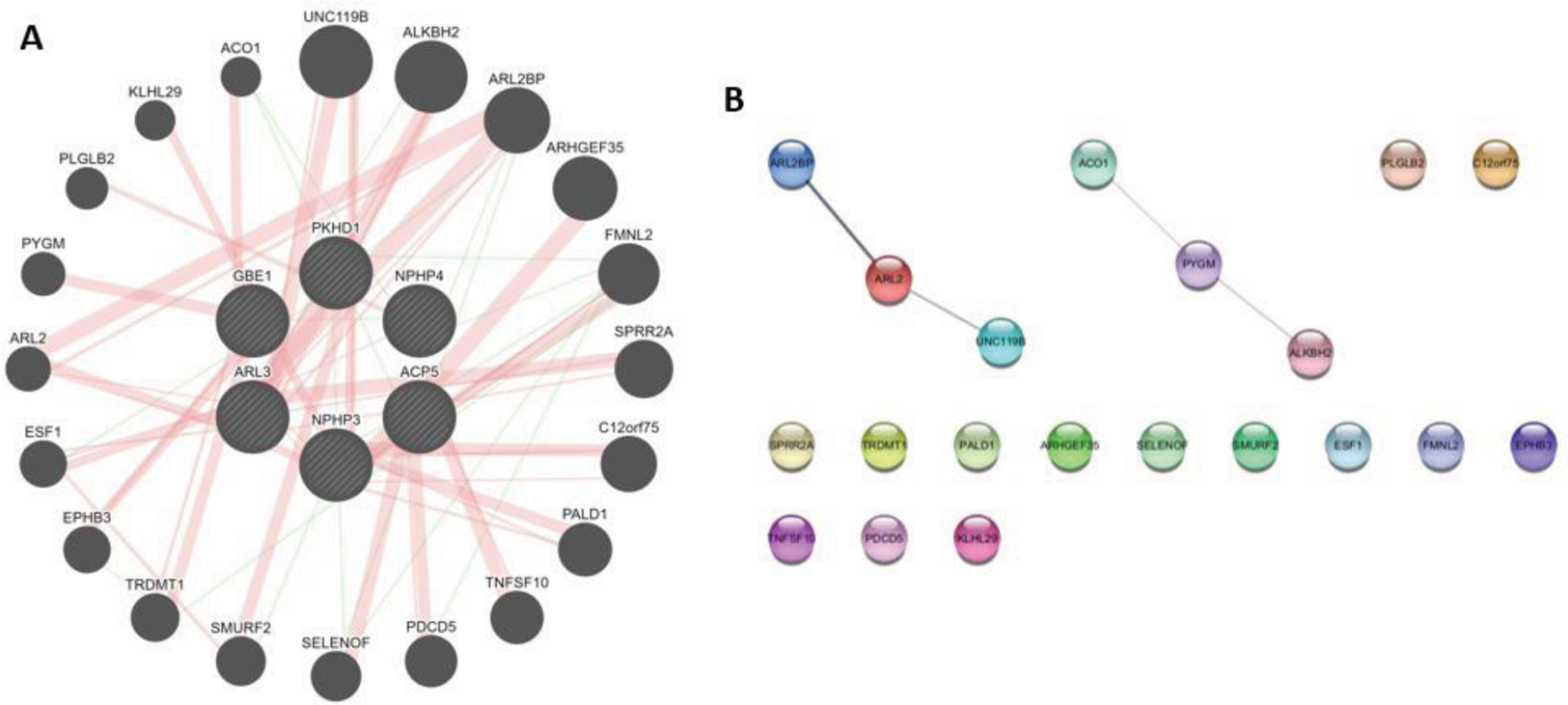
Conclusion: In this study, our results indicate that pathways of apoptosis in kidney tissues occurs along renal fibrosis in patients with LN. However, interactions among this set of genes seem to be scarce, as only 6 genes were found to interact between them. These findings warrant further research to understand the role of apoptosis in renal fibrosis secondary to LN.
REFERENCES: [1] Maria, Naomi I, and Anne Davidson. “Protecting the kidney in systemic lupus erythematosus: from diagnosis to therapy.” Nature reviews. Rheumatology vol. 16,5 (2020): 255-267. doi:10.1038/s41584-020-0401-9
[2] Hsieh, Christine et al. “Predicting outcomes of lupus nephritis with tubulointerstitial inflammation and scarring.” Arthritis care & research vol. 63,6 (2011): 865-74. doi:10.1002/acr.20441
Acknowledgements: NIL.
Disclosure of Interests: None declared.