

Background: Previous genome-wide studies have revealed >130 loci linked to systemic lupus erythematosus (SLE), primarily in European populations [1]. Research on East Asian groups has shown different genetic associations related to SLE.
Objectives: This study contributes to understanding genetic risk factors for SLE across various ancestries, focusing on the Sudanese population. This study aimed to assess genetic associations with SLE in a cohort of Sudanese individuals. We sought to determine if the HLA alleles found in this cohort align with those identified in other studies.
Methods: The study involved 483 Sudanese participants, 96 of whom were diagnosed with SLE based on the revised 1982 ACR criteria and 387 age- and sex-matched healthy controls. Genotyping was performed using the Infinium® Expanded Multi-Ethnic Genotyping Array (MEGAEX). HLA alleles and amino acids were imputed using a modified 1000G African panel in SNP2HLA [2]. Genetic markers with a minor allele frequency (MAF) below 1%, genomic missingness over 5%, or imputation quality under 75% were excluded. After quality control, 453 unrelated samples and 1,183,339 variants were analyzed statistically. Genetic associations were examined using logistic regression models adjusted for age, sex, and the first five principal components with PLINK 2.0 [3]. Significant associations were defined using Bonferroni correction for HLA alleles and amino acids, while a conventional genome-wide significance threshold was applied to other genetic associations.
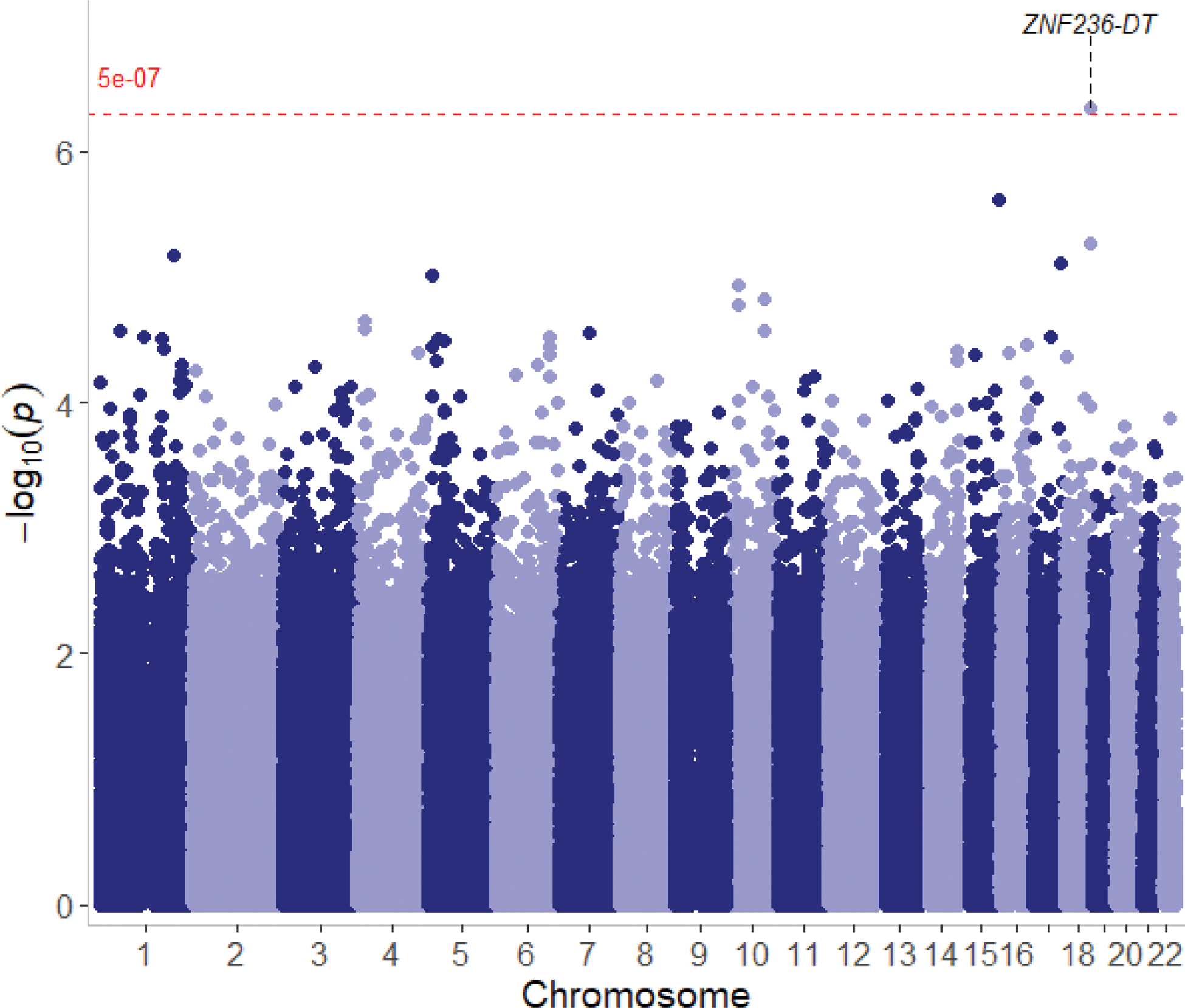
Results: The strongest association within the MHC region was found with the HLA-DRB1*03 allele (frequency = 12%), which was linked to an increased SLE risk (OR = 1.95, 95% CI = 1.19–3.16; p = 0.007). More specifically the HLA-DRB1*0301 allele was associated with a two-fold risk increase (OR = 2.00; 95% CI = 1.22–3.29; p = 0.006). Additionally, a novel association was identified with the rs12953472 marker located within the intronic region of the ZNF236-DT gene, associated with a significant increase in SLE risk (OR = 5.6, 95% CI = 2.86–10.9; p = 4.5 × 10 -07 ; Figure 1). Intriguingly, there are no prominent association signals from the MHC compared to other studies.
Conclusion: This study represents the first GWAS focused on genetic factors influencing SLE in the Sudanese population. It confirms the association of HLA-DRB1*03 with SLE and reveals a novel suggestive signal within the ZNF236-DT gene that significantly increases SLE risk. These associations need to be validated in independent studies. Future research will also focus on genetic associations to clinical manifestations of SLE in individuals of African ancestry.
REFERENCES: [1] Khunsriraksakul, C. et al. Multi-ancestry and multi-trait genome-wide association meta-analyses inform clinical risk prediction for systemic lupus erythematosus. Nat Commun 14 , 668 (2023).
[2] Jia, X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 8 , e64683 (2013).
[3] Chang, C.C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4 , 7 (2015).
Manhattan plot showing genetic associations with lupus. X and y-axes display chromosomal positions and log-transformed p-values, respectively. Genome-wide significance threshold is shown as a red dashed line.
Acknowledgements: We thank all the participants of this study.
Disclosure of Interests: Karina Patasova: None declared, NasrEldeen A Mohammed: None declared, Musa A. M. Nur: None declared, Elnour M Elagib: None declared, Amir Elshafie: None declared, Iva Gunnarsson: None declared, Elisabet Svenungsson The author declares to have inherited shares of Astra Zeneca and Pfizer, Johan Rönnelid: None declared, Sahwa Elbagir: None declared, Lina M. Diaz-Gallo: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (