

Background: Cutaneous involvement is common in Systemic Lupus Erythematosus, characterized by dysregulated immune system activation and enhanced type I interferon signaling [1, 2]. Cutaneous lupus erythematosus (CLE) encompasses three main subtypes: acute cutaneous lupus (ACLE), subacute cutaneous lupus (SCLE), and chronic cutaneous lupus (CCLE), each presenting distinct clinical outcomes and therapeutic responses [3]. The molecular differences among these subtypes and their impact on immune cell infiltration is not fully understood. Understanding the molecular and cellular landscape of each subtype can provide critical insights into their distinct pathogenic pathways and guide tailored therapeutic strategies.
Objectives: This study aimed to characterize the molecular and cellular mechanisms in the subtypes of CLE.
Methods: The dataset GSE109248 was downloaded from the Gene Expression Omnibus platform, containing transcriptomic profiles of 6 patients with CCLE, 12 with SCLE, 7 with ACLE, and 14 healthy controls (HC). Patients with CLE were compared with HC to identify differentially expressed genes (DEGs), when false discovery rate was <0.05 and log2foldchange >2. The Metascape online portal was used to conduct functional enrichment, and the Cytoscape platform to analyse protein-protein interactions. Hub genes (HGs) were identified with the Cytohubba plugin using five different algorithms. To characterize the immune cellular infiltrate, the online tool CibersortX was used, and proportions were compared with a 2-way ANOVA test while controlling the FDR. To analyse the relationship between the HGs’ expression and the immune infiltrate, a Spearman’s test was conducted. Statistical significance was set as p<0.05.
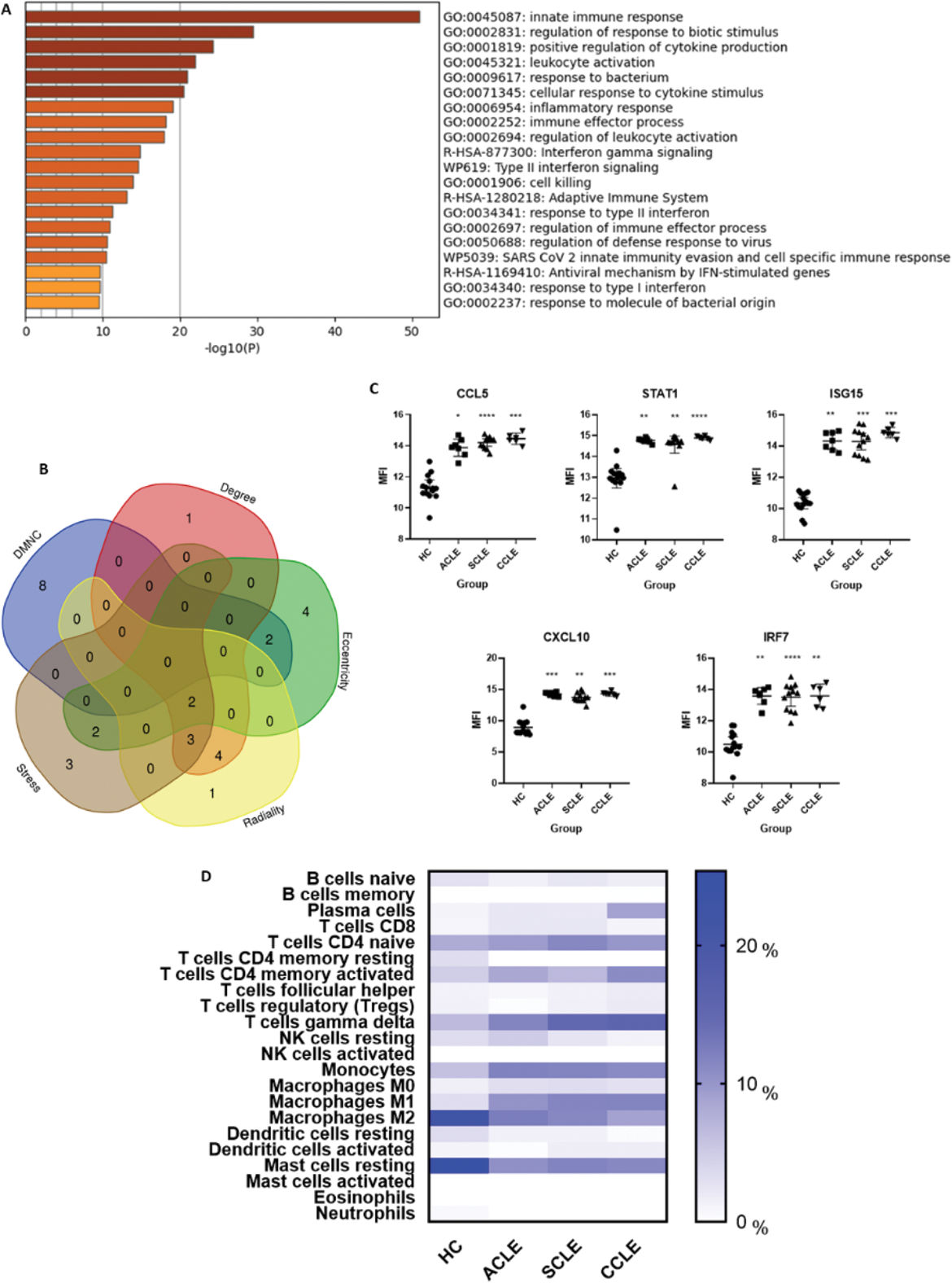
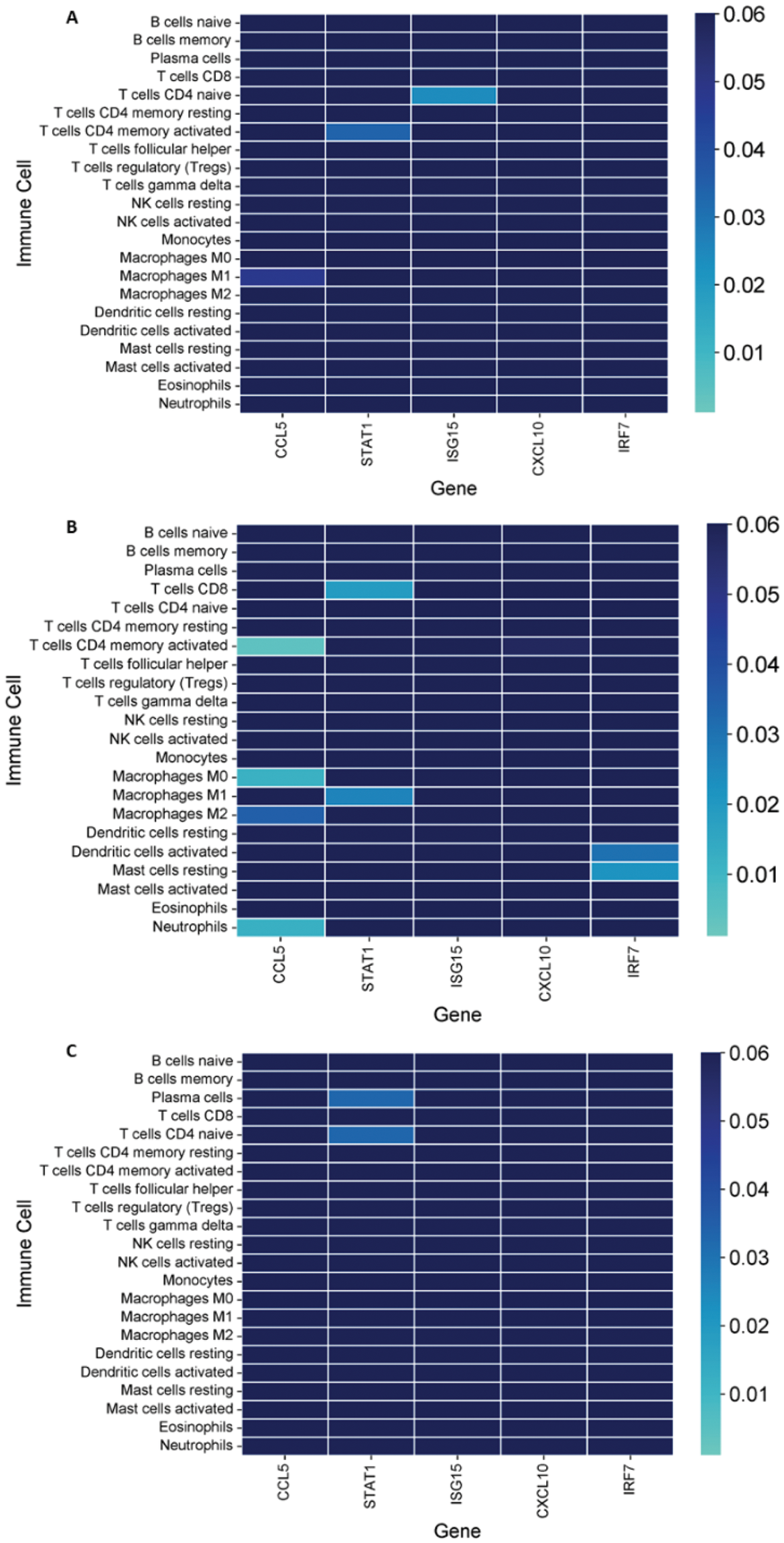
Results: 150 DEGs were identified. Functional enrichment is depicted in Figure 1A. HGs were screened, and those that overlapped in at least 3 algorithms were selected. Gene expression levels were compared retrieving the mean fluorescence intensity (MFIs) values, being significantly overexpressed in the three groups compared to HC. Significant differences were observed in the immune infiltrate of different subtypes of cutaneous lupus: plasma cells, M1 and M2 macrophages, gamma delta T cells, monocytes, and resting mast cells. Significant associations between immune cells and HGs’ expression were observed. CCL5, STAT1 and ISG15 correlated with M1 macrophages, CD4 memory activated T cells, and naïve CD4 T cells respectively (r=-0.785, 0.821, -0.857) in the ACLE group. In the SCLE group CCL5 correlated with CD4 memory activated T cells, M0 and M2 macrophages, and neutrophils (r=0.781, -0.713, -0.622, -0.707), STAT1 correlated with CD8 T cells and M1 macrophages (r=-0.676, 0.650), and IRF7 correlated with activated dendritic cells and resting mast cells (r=0.634, -0.664). In the CCLE group, STAT1 correlated to plasma cells and CD4 naïve T cells (r=-0.885, 0.887).
Functional Enrichment Analysis, Expression Levels of Key Genes, and Immune Cell Infiltration. (A ) Functional enrichment analysis of the 150 identified DEGs, showing key biological processes. (B ) Overlapping HGs identified across five different algorithms (DMNC, Degree, Eccentricity, Stress, and Radiality). (C ) Comparison of MFI values of HGs across the different subtypes of cutaneous lupus and healthy controls. The Kruskal-Wallis test with Dunn’s post-hoc comparisons was used to determine significance. Significant differences between lupus subtypes and healthy controls are indicated (*: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001, ns: not shown, p > 0.05). (D ) Heatmap showing immune cell proportions, with significant differences observed between cutaneous lupus types and healthy controls, including plasma cells, Gamma Delta T cells, monocytes, M1/M2 macrophages, and resting mast cells.
Correlation of Hub Gene Expression with Immune Cell Infiltration in Cutaneous Lupus Subtypes. (A ) Correlations between hub genes and immune cell infiltration in ACLE patients, showing significant associations particularly involving STAT1, ISG15, and CCL5 with different immune cell types. (B ) Correlations in SCLE, highlighting significant relationships between hub genes such as CCL5 and immune cell populations like macrophages and neutrophils. (C ) Correlations in CCLE patients, with STAT1 showing notable associations with plasma cells and CD4 naïve T cells. The colour gradient represents the significance, were lighter colours show higher significance.
Conclusion: The identification of distinct gene expression profiles and immune cell correlations highlights the unique pathogenic pathways involved in each subtype. Significant associations between HGs and immune cell types, such as macrophages, T cells, and dendritic cells, underscore potential targets for tailored therapeutic interventions. The observed differences in immune infiltration among ACLE, SCLE, and CCLE suggest that these subtypes may require distinct therapeutic strategies. These findings lay the groundwork for future in vivo validation and emphasize the importance of personalized approaches in the management of CLE.
REFERENCES: [1] Vale, Everton Carlos Siviero do, and Lucas Campos Garcia. “Cutaneous lupus erythematosus: a review of etiopathogenic, clinical, diagnostic and therapeutic aspects.” Anais brasileiros de dermatologia vol. 98,3 (2023): 355-372. doi:10.1016/j.abd.2022.09.005.
[2] Tsokos, G.C. “Systemic lupus erythematosus.” New England Journal of Medicine, vol. 365, no. 22, 2011, pp. 2110-2121. doi:10.1056/NEJMra1100359.
[3] Bennett, L., Palucka, A.K., Arce, E., et al. “Interferon and granulopoiesis signatures in systemic lupus erythematosus blood.” Journal of Experimental Medicine, vol. 197, no. 6, 2003, pp. 711-723. doi:10.1084/jem.20021539.
Acknowledgements: We’d like to thank the Mexican National Council of Humanities, Science and Technology (CONAHCYT) for providing the academic grant that made this research possible. Grant ID: PCC-2022-320697.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (