

Background: Sjögren’s disease (SjD) is an chronic autoimmune disorder that profoundly impacts quality of life, characterized by hallmark symptoms such as eye and oral dryness, as well as systemic manifestations including fatigue, joint pain, and organ-specific damage. Treatments remain limited. A promising way for understanding the pathophysiology of SjD lies in immunometabolism, which explores metabolic alterations within immune cells. Key findings include glucose-driven mTORC activation in human B cells, glutamine metabolism’s importance for human CD4+ T cell activation and the role of lysophosphatidic acid (LPA) in the Sjögren’s NOD mouse model [1]. To further investigate these changes, IMI PRECISESADS data [2], obtained from Sjogren’s patients, were employed for a multi-omics approach.
Objectives: Data from SjD patients within the PRECISESADS project were analyzed using the user-friendly BiomiX software [3], with a specific focus on metabolic alterations in B lymphocytes and their microenvironment. The objective was to explore multi-omics data individually and integrate them to characterize metabolic alterations and achieve a more comprehensive understanding of the disease pathology.
Methods: Bioinformatics analyses were conducted on datasets from the European PRECISESADS study (supported by the Innovative Medicines Initiative Joint Undertaking), encompassing transcriptomics, methylomics, metabolomics, and clinical data from 293 SjD patients and 508 controls. The B cell and whole blood transcriptomes were analyzed using the BiomiX transcriptomics pipeline, which included DESeq differential gene expression (DGE) analysis and metabolic pathway analysis using GSEA. Serum and urine metabolomics peaks were processed through the BiomiX metabolomics pipeline to evaluate their significance and amplitude of change. Annotation was carried out using the CEU Mass Mediator database for MS1 data (based on m/z ratio) and tidymass for MS2 fragmentation spectra. Methylomics data were analyzed with the ChAMP package integrated into BiomiX. Multi-Omics Factor Analysis (MOFA) was employed to identify shared sources of variation across the -omics datasets.
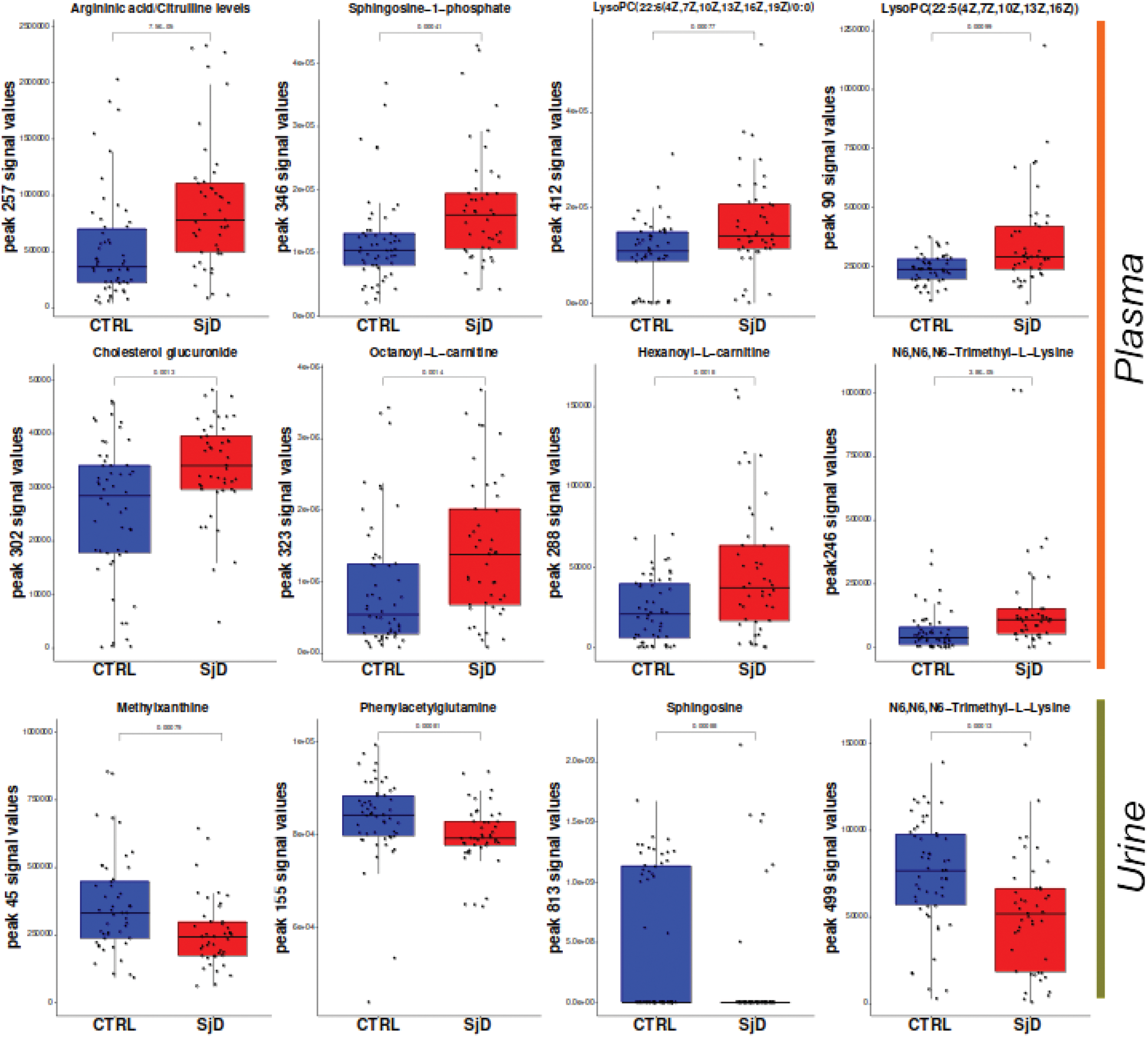
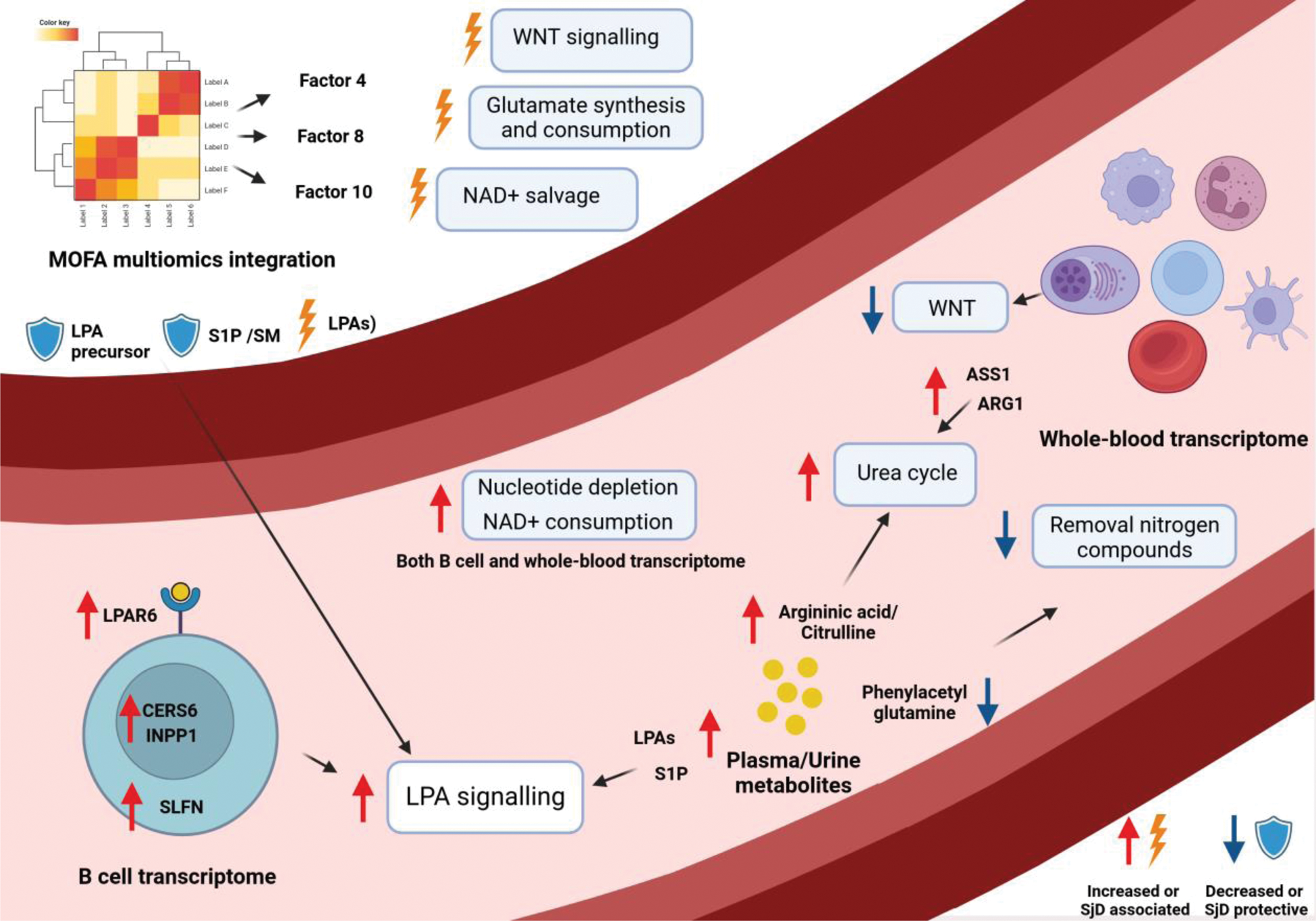
Results: Expression of LPA receptor 6 (LPAR6) in SjD B cells, along with genes involved in its metabolism as Ceramide Synthase 6 (CERS6) and Inositol Polyphosphate-1-Phosphatase (INPP1), was identified as upregulated. This finding was supported by an increase in LPA in serum and sphingosine 1-phosphate (S1P) in both serum and urine of SjD patients. The involvement of the LPA signaling pathway was further validated through MOFA analysis, which identified LPA as the primary contributor to MOFA factor 8 (linked to interferon response) and the eighth-largest contributor to factor 4 (associated with cellular activation). Conversely, S1P exhibited a protective role in MOFA factor 10, related to interferon response, alongside sphingomyelin. Additionally, increased glutamine consumption and urea cycle activity were observed. This was supported by metabolomics data (e.g., elevated argininic acid/citrulline in serum and reduced phenylacetylglutamine in urine, (Figure 1) and whole blood transcriptomic data (e.g., increased expression of Glutamine Synthetase, Peptidyl Arginine Deiminase 4, and Arginase 1 and 2). The contributions of these metabolites and genes were also linked to MOFA factor 4, suggesting an association with cellular activation. Additional metabolic alterations identified through multi-omics integration included an association of the WNT pathway association with inflammation severity and a connection between NAD+ depletion and interferon signaling. An overview of the metabolic changes is depicted in Figure 2.
Levels of metabolites in plasma and urine of control (CTRL) Sjögren’s disease patients (SjD) are depicted in blue and red, respectively. Significance was calculated using the Mann-Whitney test.
Graphical abstract illustrating intracellular signaling and metabolic pathways linked to metabolic changes in the macroenvironment from multi-omics analysis and MOFA integration. Boxes highlight altered pathways in SjD, with red or blue arrows indicating increased or decreased gene expression/metabolite levels. MOFA contributors are marked as promoting (thunderbolt) or protecting against (shield) the SjD phenotype.
Conclusion: The application of multi-omics approaches to study autoimmune diseases provides a comprehensive perspective on the underlying changes in these pathologies. In this study, metabolic alterations in SjD were investigated, highlighting a potential link between LPA and B lymphocytes via the LPAR6 receptor, as previously identified by our team in a similar study on systemic lupus erythematosus multiomics data. Additionally, we observed alterations in the B cell microenvironment, including increased glutamine consumption, activation of the urea cycle, and the involvement of the WNT pathway alterations. The consistency of these findings across multiple omics datasets strengthens our observations, proposing novel metabolic targets for understanding the pathophysiology of SjD and offering a new approach for studying autoimmune diseases.
REFERENCES: [1] Fu J et al. Pharmacological Inhibition of Glutaminase 1 Normalized the Metabolic State and CD4+ T Cell Response in Sjogren’s Syndrome. J Immunol Res .
[2] Soret P et al. A new molecular classification to drive precision treatment strategies in primary Sjögren’s syndrome. Nat Commun .
[3] Iperi C et al. BiomiX, a user-friendly bioinformatic tool for democratized analysis and integration of multiomics data. BMC Bioinformatics .
Acknowledgements: We acknowledge the 3TR and PRECISESADS consortium, which guaranteed the data used in this article and allowed us to analyze them.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (