

Background: Rheumatic diseases are highly heterogeneous, complicating stratification and personalized treatments. Clustering, an unsupervised machine learning technique, identifies meaningful subgroups by uncovering hidden patterns without relying on predefined labels.
Objectives: To review clustering techniques for identifying clinically meaningful subgroups in rheumatic diseases.
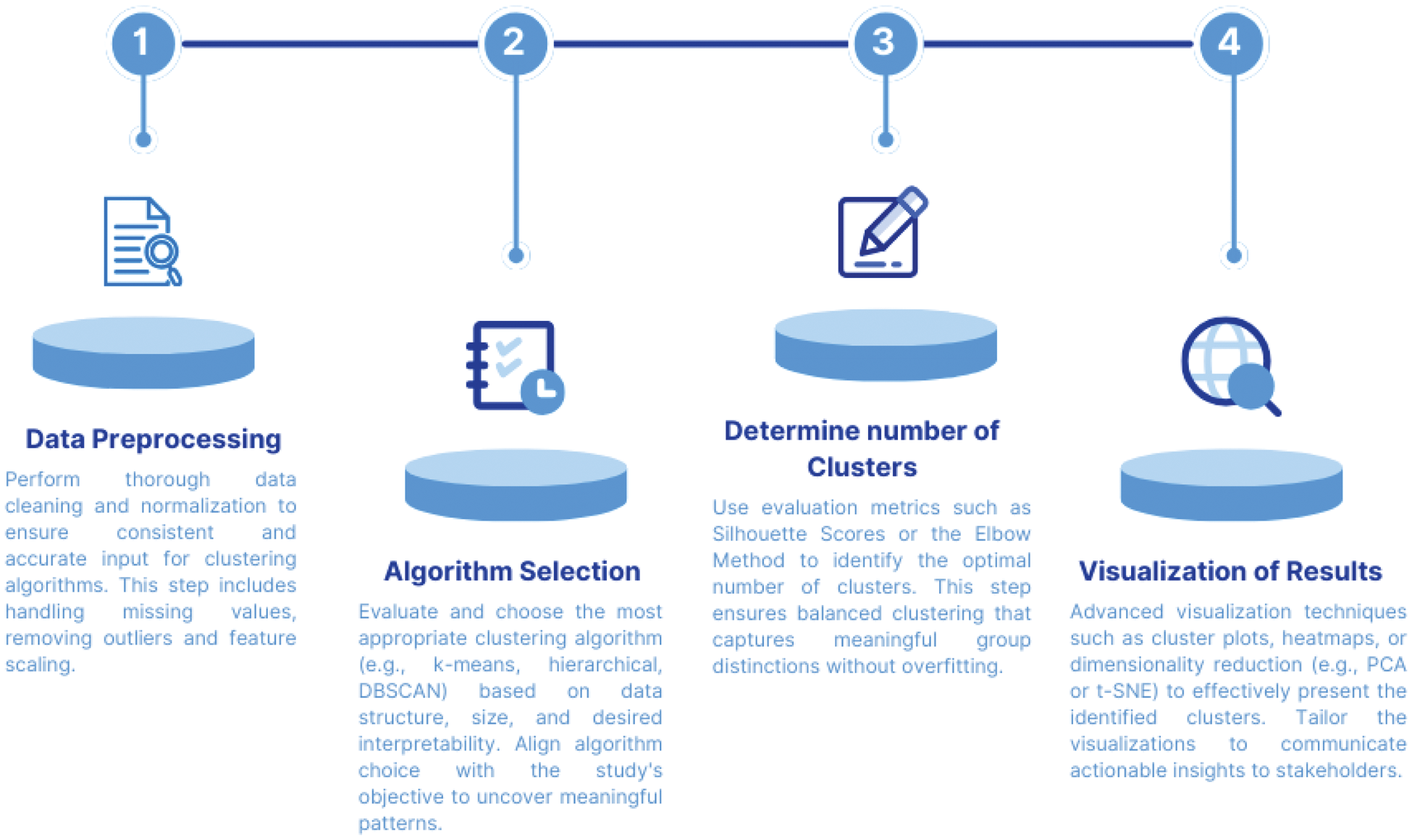
Methods: The clustering process involves sequential steps (Figure 1) to ensure robust subgroup identification. Prior to applying clustering algorithms, an Exploratory Data Analysis (EDA) was performed to evaluate the dataset’s structure, visualize data distributions, detect potential outliers, and identify relationships among key clinical variables. Proper data management is a critical foundation for reliable clustering outcomes. The first step involves data preprocessing, which is vital for the effectiveness of clustering algorithms. This includes feature scaling, to normalize variables for consistent interpretation, and management of missing data, to avoid biases that could compromise results. Following data preparation, algorithm selection tailored to clinical objectives is critical. Three methods commonly applied in rheumatology research were reviewed:
Hierarchical Clustering :
Description: builds a hierarchical tree (dendrogram) to group data points iteratively. Two main approaches are commonly used in hierarchical clustering: agglomerative (bottom-up) and divisive (top-down). In the agglomerative approach, each observation begins as its own cluster, and clusters are iteratively merged based on similarity until a single cluster containing all data points is formed. Conversely, the divisive approach starts with all data points grouped in a single cluster, which is then progressively divided into smaller clusters until each observation forms its own cluster.
Strengths: ideal for exploratory analysis; requires no predefined cluster number; provides intuitive visualizations.
Limitations: Computationally intensive for large datasets; lacks flexibility as data points cannot be reassigned after clustering.
K-means Clustering :
Description: K-means clustering partitions data into a predefined number of clusters ( k ) by iteratively assigning points to the nearest centroid and recalculating centroid positions until stabilization. To determine the optimal number of clusters, the elbow method identifies the “elbow point” in a plot of Within-Cluster Sum of Squares (WCSS) against k , where the rate of WCSS decrease slows. Alternatively, silhouette analysis evaluates clustering quality, with scores ranging from -1 to 1; values near 1 indicate well-separated clusters, near 0 suggest overlap, and negative scores imply misclassification.
Strengths: Computationally efficient; effective for well-defined, spherical clusters. Simple to implement and interpret.
Limitations: Requires prior knowledge of the number of clusters; struggles with irregularly shaped clusters and high-dimensional data.
DBSCAN (Density-Based Spatial Clustering of Applications with Noise):
Description: Identifies clusters based on density, classifying low-density points as noise or outliers.
Strengths: Does not require a predefined number of clusters; identifies irregularly shaped clusters; robust to noise.
Limitations: sensitive to parameter selection (epsilon and minPts); struggles with datasets of varying densities.
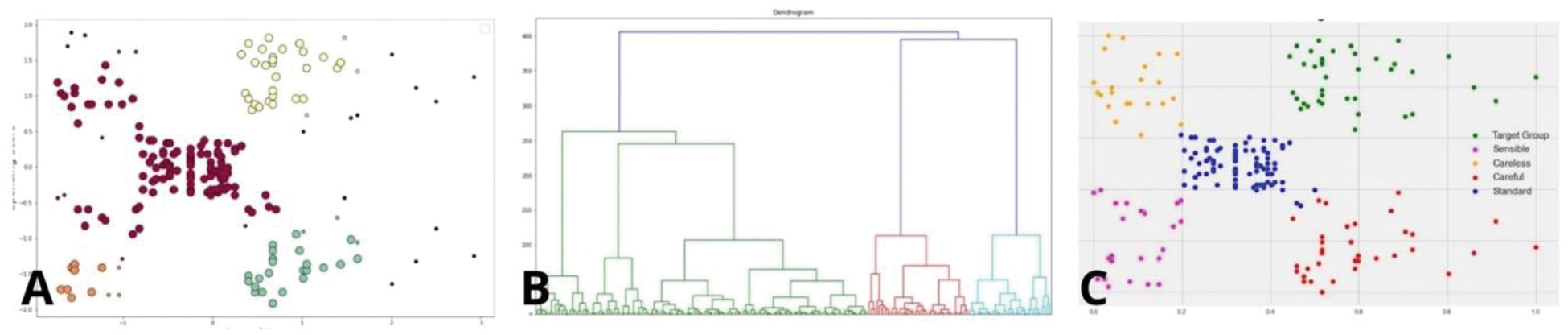
Figure 2 illustrates the distinct cluster structures generated by DBSCAN (A), hierarchical clustering (B), and K-means (C). These visualizations demonstrate how each method identifies subgroup patterns, highlighting the separation of clusters in datasets with noise (DBSCAN), hierarchical relationships (hierarchical clustering), and spherical distributions (k-means). Cluster validation involves statistical analyses to identify meaningful patterns, complemented by visualization tools like t-SNE, which simplify high-dimensional data into 2D or 3D formats. These techniques highlight cluster separation and overlap, aiding in intuitive interpretation. External cohort validation ensures the robustness and generalizability of the identified subgroups, reinforcing their clinical relevance.
Results: While this review is conceptual, prior studies validate the role of clustering in rheumatology. Clustering techniques have been applied for stratifying patients with rheumatic diseases, enabling biomarker discovery, enhancing prognosis, and optimizing treatments. There are a lot of types of clustering. However, the main thing that they share in common is the fact that they try to explain variance in the data with discrete partitions. A notable example is the study by Steinz et al. (ACR 2024), which employed pseudo-time analysis to identify and validate four distinct trajectories in early rheumatoid arthritis patients based on inflammation in blood and joints.
Conclusion: Unsupervised learning algorithms are transformative tools for phenotype and cluster identification, offering significant potential to enhance stratified care and optimize clinical trial design. By integrating clustering results into trial frameworks, researchers can ensure more precise patient selection, improving the likelihood of detecting meaningful treatment effects.
. The four continued steps in the clustering analysis workflow.
. Illustrative examples of clustering results produced by DBSCAN (A), hierarchical clustering (B), and K-means (C). These examples are general representations intended to visualize algorithm outputs and are not based on the data analysis.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: Asier García-Alija Amgen, Berta Paula Magallares GSK, AstraZeneca, Hye Sang Park Lilly, Pfizer, Alfa Sigma, Janssen, Novartis, MSD, Boeringer and Amgen, Guillem Verdaguer: None declared, Ana Laiz Abbvie, Johnson&Johnson, UCB, Novartis, UCB, Patricia Moya GSK, Albert Casals Urquiza: None declared, Cesar Díaz-Torné Abbvie, Lilly, Alfasigma, Novartis, UCB, Luís Sainz Comas Johnson&Johnson, Abbvie, Lilly, Alfasigma, Novartis, UCB, Ivan Castellví Boehringer-Ingelheim, GSK, Novartis, Boehringer-Ingelheim, GSK, Johnson&Johnson, Novartis, Boehringer-Ingelheim, Sanofi, GSK, Susana P. Fernandez-Sanchez GSK, AstraZeneca, Julia Bernardez Otsuka, Helena Codes: None declared, Jose Luis Tandaipan Johnson&Johnson, Concepción Pitarch: None declared, CARLA MARCO PASCUAL: None declared, Andrea Garcia Guillen: None declared, Núria Fernández-Verdés: None declared, Lorena Úbeda: None declared, Maria Àngels Melchor: None declared., Margarita Sihuro: None declared, Sandra Ros: None declared, Hèctor Corominas: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (