

Background: Systemic lupus erythematosus (SLE) exhibits considerable heterogeneity in disease severity and progression, complicating its study and management. The SLE Disease Activity Index (SLEDAI) is a critical tool for assessing disease activity, but its variability can affect clinical trial outcomes, potentially influencing patient recruitment. Screening failures in clinical trials due to unmet inclusion criteria are a significant challenge, with a reported failure rate of approximately 15%.
Objectives: The goal of this study is to better understand the dynamics of SLEDAI variability and explore ways to optimize patient recruitment and trial design. Specifically, we aim to analyse the variability of individual SLEDAI components over time, identifying those that exhibit stable progression and those with recurrent patterns across different patients.
Methods: We utilized a comprehensive patient database from TransCelerate BioPharma Inc., incorporating data from 8 biopharmaceutical companies, including shared placebo arms. A harmonization tool powered by large language models (LLMs) was developed to integrate and ensure the consistency and accuracy of diverse clinical trial data. Using this data, we assessed the importance of SLEDAI score components and their contribution to patient condition.
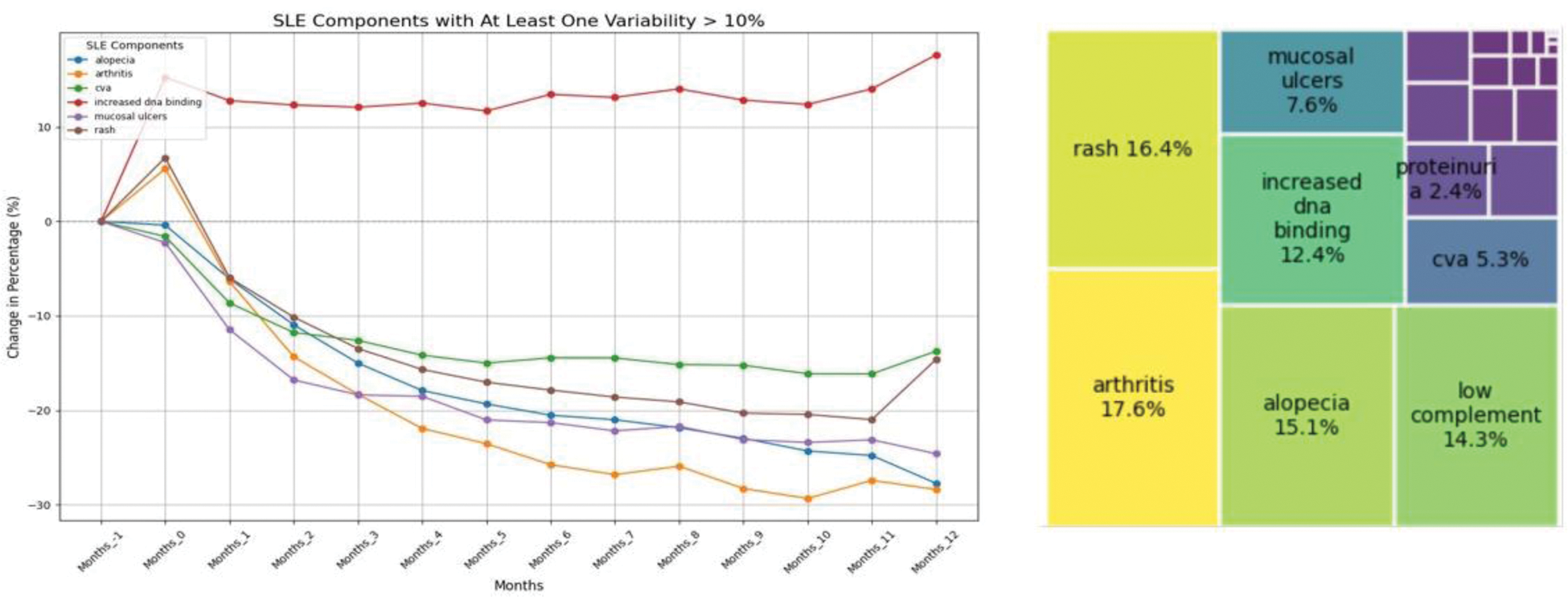
Results: Analysis of data from 2,649 patients across 12 clinical trials revealed valuable insights into the variability and contribution of individual SLEDAI components over time. By examining each component, we identified those that contributed to score instability during trial periods. Furthermore, we quantified the contribution of each component to the overall SLEDAI-2K score, emphasizing on their average percentage contributions to the final score. Our findings (Figure 1) revealed:
High Contribution (>10%) & High Variability (>10% Change): Components such as Arthritis, Alopecia, Rash, and Increased DNA Binding showed significant variability and contributed substantially to score instability.
High Contribution (>10%) & Low Variability (≤10% Change): Low Complement was found to have a stable variability but a strong contribution to the final score.
These findings contribute to a deeper understanding of SLEDAI dynamics, with implications for improving trial design and refining patient selection. Developing predictive machine learning models for disease activity, targeting individual components or their combinations, can identify key baseline characteristics, enhance recruitment strategies, and advance disease understanding.
Conclusion: Understanding the variability of individual SLEDAI components and their role in disease progression provides valuable insights for reliably assessing treatment efficacy in clinical trials. This analysis can support refining trial design through a better understanding of disease activity patterns, which may lead to identifying the right patients for recruitment and achieving more effective study outcomes.
Left, percentage change in the presence of specific components of the SLEDAI score over the duration of the clinical trial, relative to pre-screening (1 month before the trial start). Only components with an absolute variability greater than 10% over time are shown. The analysis highlights that the components driving the greatest variability in the final score include alopecia, arthritis, cva, increased dna binding, mucosal ulcers, and rash. Right: Contribution of each SLEDAI score component to the final score. Components with a higher contribution to the overall score include arthritis, rash, alopecia, increasing dna binding and low complement. Low complement, despite showing minimal variability, exhibits a significant impact on the final score.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: Caterina Tozzi AstraZeneca, Adam Taylor AstraZeneca, Claire Donoghue AstraZeneca, SARAH HOTEE AstraZeneca, Gabriel Wong AstraZeneca, Starr chen AstraZeneca.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (