

Background: Circulating monocytes are the precursors of tissue-infiltrating macrophages, which serve as critical immune mediators in rheumatoid arthritis (RA). Understanding early metabolic changes in circulating monocytes is of crucial importance for the understanding of disease mechanisms [1]. This project focuses on tryptophan metabolism, particularly the kynurenine pathway, in monocytes (Figure 1A), which has been previously reported to be altered in RA and was linked to inflammation; however, the specific regulation remains unclear [2].
Objectives: This study aimed to elucidate regulation of tryptophan metabolism in circulating monocytes of RA patients and to compare them to healthy donors (HDs).
Methods: CD14+ monocytes (positive selection with CD14 Microbeads) were isolated from PBMCs of sero-positive RA patients and age and sex matched HDs. The cells were cultured for 24 hours and stimulated with vehicle or 100 ng/mL lipopolysaccharide (LPS). Metabolites were extracted using 50% methanol, 30% acetonitrile and 20% ultrapure water using Liquid Chromatography-Tandem Mass Spectrometry (LC MS/MS) method. The peak area of each detected metabolite was normalized against the total ion count and statistically analyzed using two-way ANOVA with Šídák multiple comparison in GraphPad PRISM. Bulk RNA transcripts were analyzed with the DeSeq2 package followed by gene set enrichment analysis (GSEA) using the tryptophan metabolism gene list from KEGG. Proteins were extracted with SDS containing lysis buffer and acquired by LC MS/MS method. Protein abundances were transformed using the variance stabilizing normalization and further statistically analyzed as described for metabolomics above.
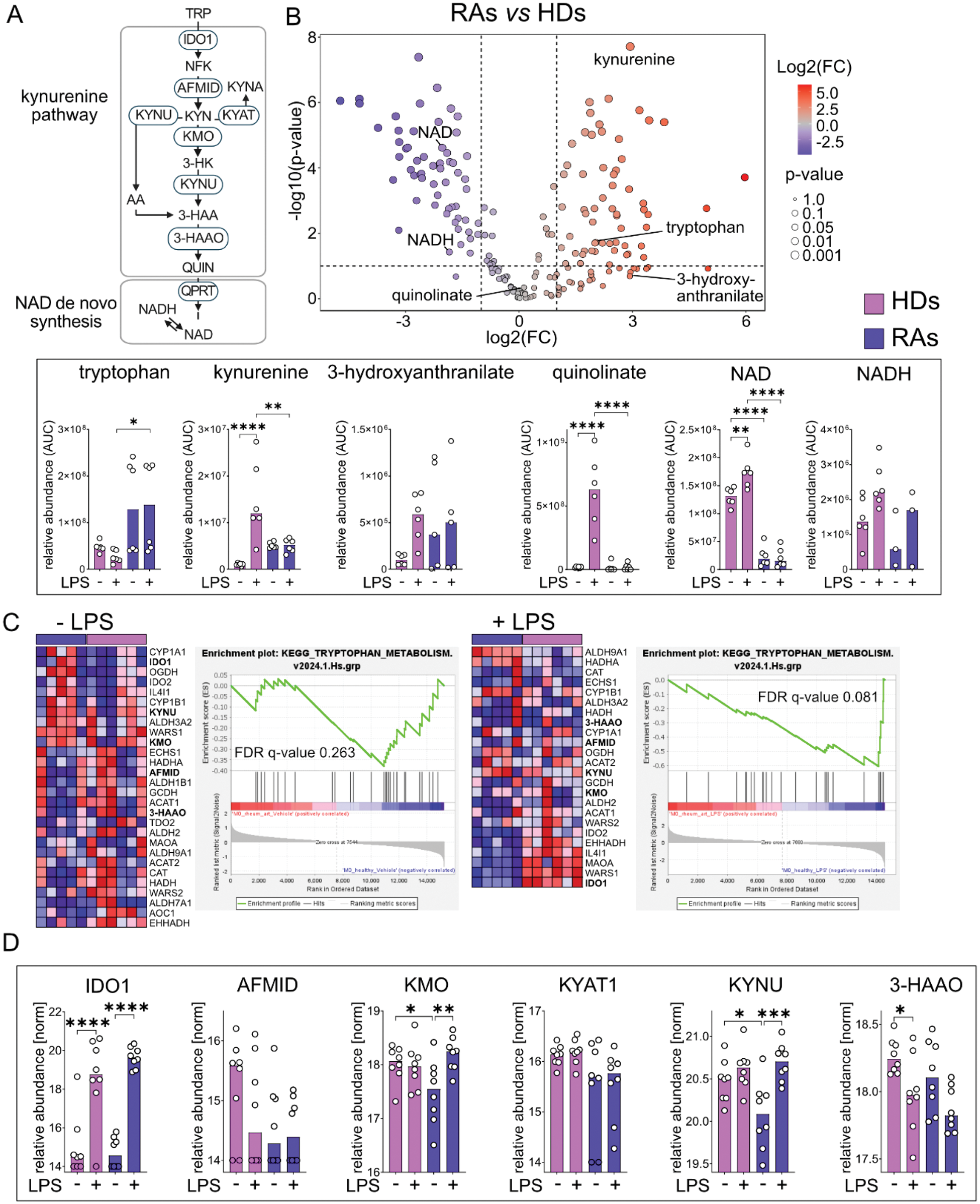
Results: Metabolomic analyses revealed upregulation of the proximal kynurenine pathway, which demonstrates one side branch of the tryptophan (TRP) metabolism, in RA patients with increased levels of kynurenine (KYN) as well as a 2-fold increase of 3-hydroxyanthranilate (3-HAA), whereas downstream metabolites such as quinolinate (QUIN) remain unchanged and we observed a reduction of nicotinamide adenine dinucleotide (NAD) in RAs under basal conditions. Upon LPS stimulation, monocytes from HDs demonstrated upregulation of kynurenine, quinolinate, NAD and 1.5-fold increase of 3-hydroxyanthranilate which represents the expected dynamics of tryptophan metabolism during inflammatory stimulation. Conversely, RA patient-derived monocytes showed a reduced inflammatory response (Figure 1B).
Transcriptomic analyses revealed no clustering of RA patients and HDs under basal conditions yet supported metabolic observations through elevated expression of Indoleamine 2,3-Dioxygenase 1 ( IDO1 ) within the proximal kynurenine pathway. Notably, LPS treatment led to a clustering between RA patients and HDs, showing a higher expression of IDO1 in HDs. Furthermore, GSEA demonstrated under LPS treatment an overall downregulated tryptophan metabolic profile in RA, consistent with previous metabolomic observations (Figure 1C).
Proteomics showed that in RA patients there is no enzymatic dysregulation of proteins involved in tryptophan metabolism after LPS stimulation. Both RA patients and HDs showed efficient upregulation of IDO1 in response to LPS. Notably, lower Kynurenine 3-Monooxygenase (KMO) and Kynureninase (KYNU) levels were observed in RA patients compared to HDs on basal conditions (Figure 1D).
Conclusion: Under basal conditions, proximal metabolites of the kynurenine pathway were increased in monocytes from RA patients compared to HDs, while distal metabolites remained unchanged or decreased. However, RA patients showed impaired upregulation of tryptophan metabolism after addition of LPS compared to healthy individuals.
REFERENCES: [1] Weyand, C. M., Zeisbrich, M., & Goronzy, J. J. (2017). Metabolic signatures of T-cells and macrophages in rheumatoid arthritis. Current opinion in immunology, 46, 112–120.
[2] Moulin, D., Millard, M., Taïeb, M., Michaudel, C., Aucouturier, A., Lefèvre, A., Bermúdez-Humarán, L. G., Langella, P., Sereme, Y., Wanherdrick, K., Gautam, P., Mariette, X., Dieudé, P., Gottenberg, J. E., Jouzeau, J. Y., Skurnik, D., Emond, P., Mulleman, D., Sellam, J., & Sokol, H. (2024). Counteracting tryptophan metabolism alterations as a new therapeutic strategy for rheumatoid arthritis. Annals of the rheumatic diseases, 83(3), 312–323.
(A ) Scheme of tryptophan metabolism with the kynurenine pathway and NAD de novo synthesis. (B ) Volcano plot of untargeted metabolomics from monocytes of rheumatoid arthritis patients (RAs) compared to healthy donors (HDs) under basal conditions (N=6). Corresponding bar plots show medians of tryptophan metabolites with their relative abundance as area under the curve (AUC) on the y-axis. (C ) Gene set enrichment analysis with corresponding heatmap comparing HDs (N=6) with RAs (N=5) under basal condition and after LPS treatment. (D ) Bar plots showing the medians of tryptophan-associated proteins with normalized values of the relative abundance on the y-axis (N=6). Imputed proteomics values are designated as filled dots. Statistical significance was tested using a two-way ANOVA test. P-values signify *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001.
Acknowledgements: This project was funded by the Swiss National Science Foundation (SNSF) and we would like to thank all participating patients from Basel and Zurich.
Disclosure of Interests: Silja Vittoria Malkewitz: None declared, Seyram Maureen Duphey: None declared, Celia Makowiec: None declared, Ming Yang: None declared, Christian Frezza: None declared, Diego Kyburz Abbvie, UCB, Sanofi, Novartis, Pfizer, Roche, Jessica Jäger: None declared, Stefan Dudli: None declared, Oliver Distler has/had consultancy relationships with and/or has served as a speaker for the following companies in the area of potential treatments for systemic sclerosis and its complications in the last three calendar years: 4P-Pharma, Abbvie, Acceleron, Acepodia Biotech, Aera, Alcimed, Altavant, Amgen, AnaMar, Anaveon AG, Argenx, AstraZeneca, Blade, Bayer, Boehringer Ingelheim, Calluna (Arxx), Cantargia AB, Catalyze Capital, Corbus, CSL Behring, Galderma, Galapagos, Glenmark, Gossamer, Horizon, Janssen, Kymera, Lupin, Medscape, MSD Merck, Miltenyi Biotec, Mitsubishi Tanabe, Nkarta Inc., Novartis, Orion, Pilan, Prometheus, Quell, Redxpharma, Roivant, EMD Serono, Topadur and UCB, Patent issued “mir-29 for the treatment of systemic sclerosis” (US8247389, EP2331143).Co-founder of CITUS AG, Research Grants: BI, Kymera, Mitsubishi Tanabe, UCB, Bojana Müller receives funding from the Swiss Science Foundation, Goldscmidt-Jacobson Foundation, Novartis Foundation for Biological-Medical Research, Iten-Kohaut Foundation and UniScientia Foundation. Conference support is provided by Johnson&Johnson.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (