

Background: As part of the Accelerating Medicines Partnership (AMP), we discovered that urinary PR3, a neutrophil degranulation product, is linked to histological activity in proliferative lupus nephritis (LN) and is detectable in LN biopsies. Because mature neutrophils are not frequently detected by optical microscopy in LN kidney biopsies, we hypothesized that PR3 derived from other cell types may be implicated in LN pathogenesis.
Objectives: To investigate the association of intrarenal PR3+ cells with clinical and pathological features, and to characterize their immunophenotype.
Methods: We conducted multiplexed histology on LN biopsies using serial immunohistochemistry (sIHC) to quantify PR3 expression along with multiple cell lineage markers (20-plex). Image analysis, including deconvolution, cell segmentation and quantitative histology, was performed with IndicaLab HALO. We identified PR3+ cells with a fluorescence threshold, then used a 20-marker signature to exclude false positives, and applied 10 markers to specifically define subclusters. Spatial transcriptomic analyses were conducted using Nanostring GeoMX (near transcriptome-wide RNA profiling) on one kidney biopsy. PR3+ cells were defined by immunofluorescence based on PR3+ and nuclear staining (SYTO). RNA profiles were cross-referenced with publicly available transcriptional signatures of circulating myeloid cells as well as kidney-infiltrating immune cells [1-3].
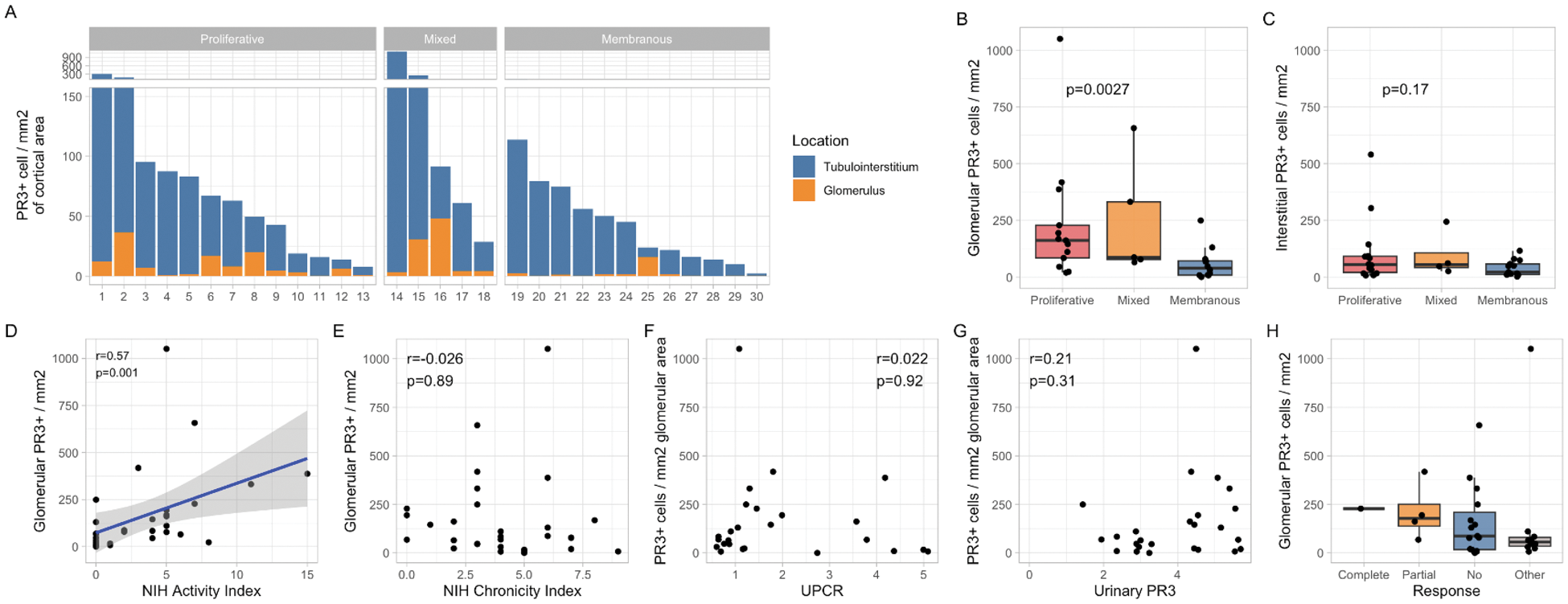
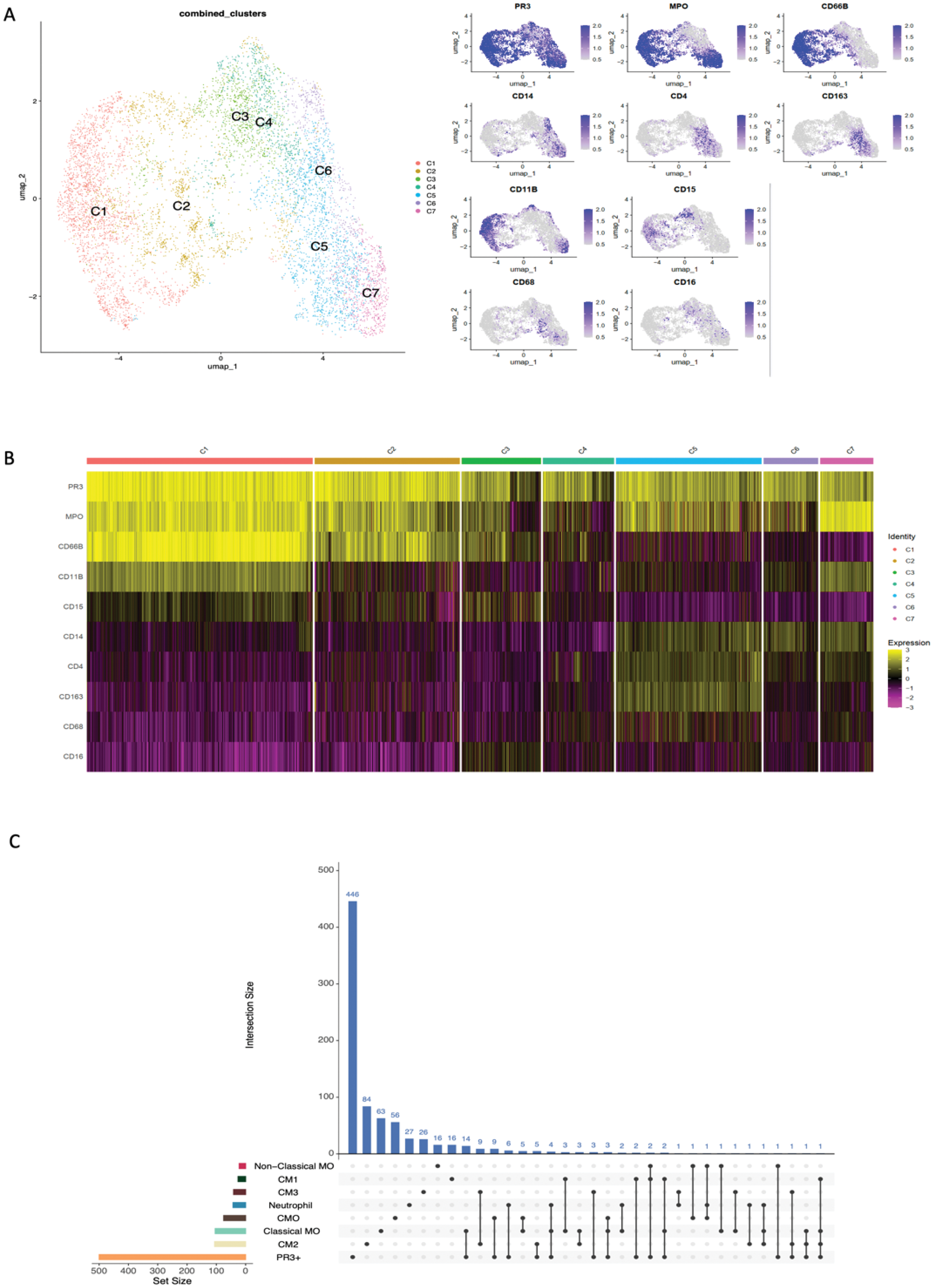
Results: A total of 33 patients with LN were enrolled: 14 (42.4%) with pure proliferative LN, 14 (42.4%) with pure membranous LN and 5 (15.2 %) with a mixed phenotype. PR3+ cells were present in all LN biopsies, predominantly in the tubulointerstitium (Figure 1A). Patients with proliferative LN had significantly higher glomerular PR3+ cell density compared to membranous LN (p=0.0037, Figure 1B). Glomerular PR3+ density correlated with histological activity (NIH Activity Index, r=0.57, p=0.001) but not with the Chronicity Index or proteinuria (Figure 2D-F ). Urinary PR3+ levels did not correlate linearly with intrarenal PR3+ density, though higher urinary PR3+ tended to align with increased intrarenal density (Figure 1G ). We could not definitively study the association between PR3+ density and response given the limited number of responders (Figure 1H ). Tissue proteomic analysis identified seven distinct subsets of PR3+ cells based on the co-expression of various lineage-specific markers (Figure 2A,B ). Myeloperoxidase (MPO) was coexpressed in most clusters. A group of clusters including C1-3, expressed markers typical of classical neutrophils (CD66b and CD15). A second group including clusters C5-7 showed high CD14 expression suggesting monocyte/macrophage lineage and included one cluster (C5) with macrophage/phagocytes markers like CD163 and CD68. Spatial transcriptomic analysis revealed that transcriptional features of PR3+ cells had the largest overlap with classical monocytes, inflammatory CD16+ (CM0) or phagocytic (CM1) macrophages, and neutrophils, supporting the heterogeneity of PR3+ cells (Figure 2C ).
Conclusion: PR3+ cells are abundant in LN, increased in proliferative classes, and strongly associated with histological activity, characterizing a more aggressive disease phenotype and implicating PR3 degranulation in LN pathogenesis. PR3 is expressed by several cell types within the lineage of neutrophils and monocytes/macrophages. These findings support the potential role of targeting these cells in LN.
REFERENCES: [1] Hong Yet al. iScience. 2022.
[2] Villani AC et al. Science. 2017.
[3] Arazi A et al. Nat Immunol. 2019.
Intrarenal PR3+ Cell Distribution and Associations with Clinical Features in Lupus Nephritis Patients
(A ) Distribution of PR3+ cell density in the glomerular and tubulointerstitial areas according to histological class and location. (B-C ) Box plots comparing glomerular and tubulointerstitial PR3+ cell density across LN classes. The Correlation between PR3+ glomerular cell density and NIH Activity and Chronicity indices is reported in panels D and E . Panels F and G show the association between glomerular PR3 cell density and UPCR and urinary PR3, respectively. Panel H display glomerular PR3+ density according to clinical response (but only 1 case of Complete Response.
Phenotypic Characterization of PR3+ Cells based on proteomic (A,B) and transcriptional profiles (C).
(A ) UMAP plot illustrates the seven distinct cell clusters (C1-C7), labeled and distinguished by color. On the right, UMAP displaying the sIHC expression levels of PR3, MPO, and various lineage markers (CD66b, CD11b, CD15, CD14, CD4, CD163, CD68, CD16) across the distinct PR3+ cell clusters. (B ) Heatmap shows the sHIC expression level of the previous mentioned markers. Each column is a single PR3+ cell and each row is a single marker. Yellow indicates higher expression, and purple indicates lower expression, highlighting the phenotypic diversity of PR3+ cells in LN kidney biopsies. (C )UpSet plot showing the intersection between PR3+ cells transcriptional pattern and other cell populations (sets) gene expression profiles, including classical monocyte (MO), nonclassical monocyte, neutrophil, CM0 (Inflammatory CD16+ macrophages), CM1 (Phagocytic CD16+ macrophages), CM2 (tissue-resident macrophages) and CM3 (conventional dendritic cells). The horizontal colored bars on the left represent the size of each set. The vertical blue bars indicate the size of the intersections between sets. For example, there were 14 genes uniquely shared between intrarenal PR3+ cells and classical monocytes (in addition to several others also expressed by other myeloid cells).
Acknowledgements: NIL.
Disclosure of Interests: Alessandra Ida Celia: None declared, Chen-Yu Lee: None declared, Taibo Li: None declared, Eugene Shenderov: None declared, Kelly Casella: None declared, Angelique Cortez: None declared, Sarah Louis: None declared, Xiaoping Yang: None declared, Cristiano Alessandri: None declared, Fabrizio Conti: None declared, Michelle Petri AbbVie, Alexion, Amgen, AnaptysBio, Annexon Bio, Arthros-FocusMedEd, AstraZeneca, Atara Biosciences, Aurinia, Autolus, Bain Capital, Baobab Therapeutics, Biocryst, Biogen, Boxer Capital, Cabaletta Bio, Caribou Biosciences, CTI Clinical Trial and Consulting Services, CVS Health, DualityBio, Escient Pharmaceuticals, Emergent Biosolutions, Exo Therapeutics, Gentibio, GSK, iCell Gene Therapeutics, IconPLC, IQVIA, Innovaderm Research, Kezar Life Sciences, Kira Pharmaceuticals, Eli Lilly, Merck EMD Serono, Nexstone Immunology, Nimbus Lakshmi, Novartis, Ono Pharma, Owkin, PPD Development, Proviant, Regeneron, Seismic Therapeutic, Senti Biosciences, Sinomab Biosciences, Steritas, Takeda, Tenet Medicines, TG Therapeutics, UCB, Variant Bio, Worldwide Clinical Trials, Zydus, Aurinia, Exagen, GSK, Janssen, the Accelerating Medicines Partnership in RA/SLE Network: None declared, Sladjana Skopelja-Gardner: None declared, Avi Rosenberg: None declared, Andrea Fava AstraZeneca, BMS, Artiva, AnnexonBio, Sanofi, UCB, BAin Capital, Novartis, Exagen.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (