

Background: Some cases of systemic lupus erythematosus (SLE) are resistant to treatment, and patients suffer serious effects such as progression of organ damage and increased adverse effects of treatment. Treatment resistance is a major barrier to achieving remission or low disease activity. To resolve the issue, research has been conducted on factors related to treatment resistance, including genetic analysis. Analysis of the relationship between changes in gene expression before and after treatment and treatment response, mainly focusing on cell-type-specific activity signatures, has been reported [1]. However, there are few reports on the analysis of treatment resistance that take the passage of time into account by performing time-course analysis using longitudinal PBMC samples.
Objectives: This study aimed to elucidate the factors that cause treatment resistance in SLE in clinical practice using RNA sequencing analysis of longitudinal PBMC samples. We extracted differentially expressed genes (DEGs) between 12 weeks after treatment and before treatment and confirmed the differences between the treatment responders and treatment-resistant group. We then performed time-course analysis using samples before treatment and 4 and 12 weeks after treatment to identify critical factors that are involved in treatment resistance over time, considering long-term changes.
Methods: Thirty-nine peripheral blood samples were collected over time from naïve or flare SLE patients who received standard induction therapy except for belimumab, anifrolumab, and rituximab. PBMCs were collected before treatment and 4 and 12 weeks after treatment. Patients were divided into two groups based on their response to treatment: those who achieved a SLEDAI-2K score of less than 4 after 12 weeks as the treatment responder group, and those who did not achieve this score as the treatment-resistant group. RNA sequencing was performed using NovaSeq 6000 (Illumina) with a paired-end read of 100 base pairs. FASTQ format data were mapped to the UCSC human genome 38 as the reference sequence. Sequence read count data was analyzed using R (4.3.2). DEGs were identified using EdgeR. Time-course expression analysis was performed using maSigPro.
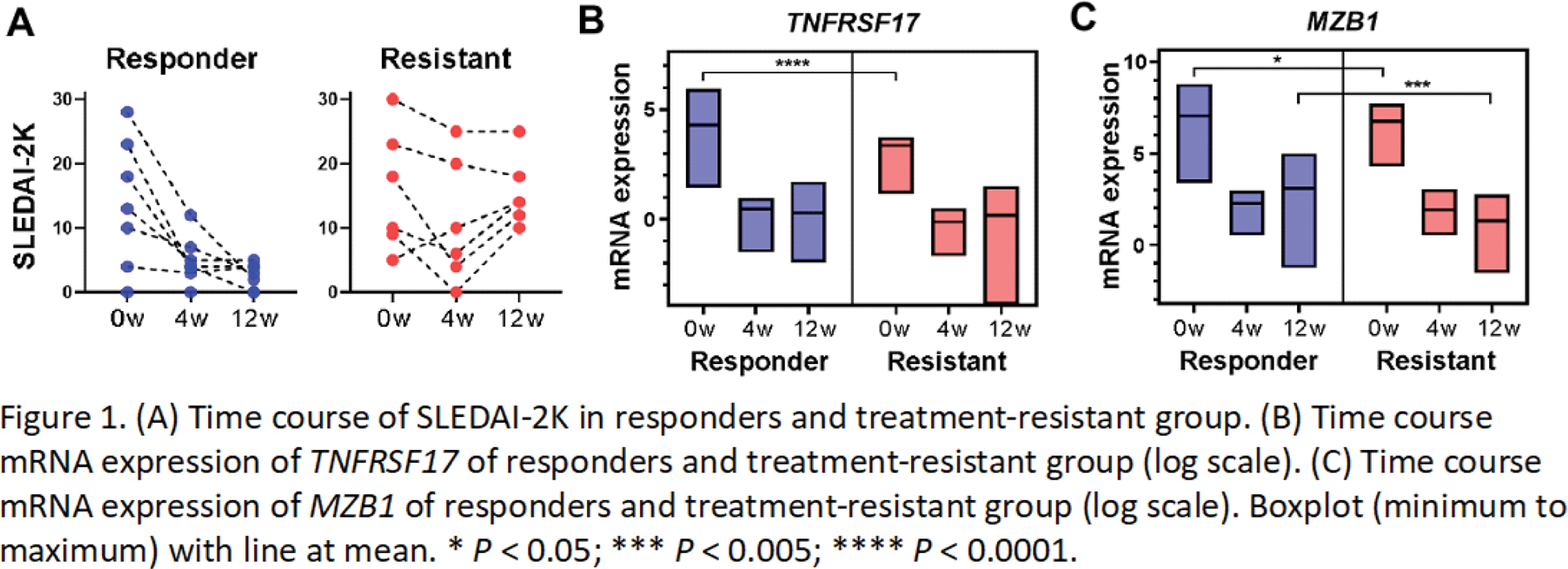
Results: Of the 13 patients who received standard induction therapy, seven responded to treatment and six were resistant (Figure 1A). The number of DEGs comparing before treatment and 12 weeks after treatment was higher in the responder group (1813 in the responder group, 366 in the treatment-resistant group, and 163 genes common to both groups). Many of the 203 genes found only in the treatment-resistant group were related to IFN signature genes. Time-course analysis was performed using PBMC samples before treatment and 4 and 12 weeks after treatment. The treatment responder and treatment-resistant group were compared using maSigPro, and the differentially expressed genes in the time series were extracted. In addition to immunoglobulin-related genes, TNFRSF17 ( P = 6.7021 x 10 -11 ) (Figure 1B) and MZB1 ( P = 3.5796 x 10 -11 ) (Figure 1C) were identified. Both genes were elevated in the responders and significantly lower in the treatment-resistant patients before treatment, and then declined in essentially the same way after treatment intervention. TNFRSF17 (BCMA), a receptor for BAFF/BLyS, has been investigated as a therapeutic target for SLE [2]. MZB1 is a B-cell-specific ER chaperone complex that plays a key role in antibody secretion and is increased in SLE lymph nodes [3]. These molecules may be critical for treatment resistance.
Conclusion: Time-course analysis using longitudinal PBMC samples suggested that the expression levels and trends of TNFRSF17 and MZB1 are important for treatment resistance in SLE.
REFERENCES: [1] Nakano M, Ota M, Ishigaki K, Fujio K, et al. Distinct transcriptome architectures underlying lupus establishment and exacerbation. Cell. 2022;185(18):3375-3389.e21.
[2] Wang W, He S, Yuan Y, et al. BCMA-CD19 compound CAR T cells for systemic lupus erythematosus: a phase 1 open-label clinical trial. Ann Rheum Dis. 2024;83(10):1304-1314.
[3] Miyagawa-Hayashino A, Yoshifuji H, Tsuruyama T, et al. Increase of MZB1 in B cells in systemic lupus erythematosus: proteomic analysis of biopsied lymph nodes. Arthritis Res Ther. 2018;20(1):13.
Acknowledgements: NIL.
Disclosure of Interests: Shuji Sumitomo Asahi-kasei, GSK, AstaraZeneca, Abbvie, Pfizer, Eli Lilly, Chugai, Astellas, BMS, Eizai, Mitsubishi-Tanabe, Hideki Oka: None declared, Takeshi Iwasaki: None declared, Koichiro Ohmura Eisai, Gilead Sciences, AstraZeneca, GSK, Asahi-Kasei Pharma, Chugai Pharmaceutical, Mitsubishi Tanabe Pharma.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (